Transcriptional Rhythm
Seth Darst
Rockefeller University
Published January 27, 2018
If the rhythm of bacterial genes being transcribed was set to music, it might sound like the asymmetric beat of Bartok, a favorite composer of structural biologist and pianist Seth Darst.
In bacteria, the core transcription enzyme making RNA from the DNA template rattles off about 20 nucleotides a second. But every 10 seconds or so, the swift RNA transcription pauses. Transcribing RNA from a gene may take 1-2 minutes, with half of that time spent on hold. The pauses happen at predictable intervals and are presumed to be important, but so far their function remains mostly a mystery.
In his laboratory and at the piano keyboard, Darst aims to decipher the rhythms in science and sonata. At the Rockefeller University in New York, Darst studies RNA transcription in bacteria to understand the details. After all, making RNA from the DNA template is the first step in gene expression in all organisms.
In the last two decades, the Darst lab has been building a portfolio of atomic images that he and others once thought impossible in their lifetimes. In fact, the work Darst does today might have surprised his earlier self, who was on track to become a physician or a professional musician before a lecture in a required undergraduate biochemistry class sowed the seeds for his scientific future.
Darst grew up in Seattle, where his father built houses. In elementary school, he preferred to read books about the solar system or the ocean instead of recess. An economic downturn prompted a family move to Loveland, Colorado, on the plains at the base of the Rocky Mountains. Darst finished high school a year early and helped his father in his homebuilding business. Darst was an early student of his piano-teacher mother. After considering a career in music, he enrolled in biochemical engineering at the University of Colorado, Boulder. The premed program simultaneously met the expectation he would follow in the footsteps of his physician grandfather and kept his future options open. Darst remembers being “totally blown away” by a lecture in structural biology in an introductory biochemistry class taught by RNA biologists Larry Gold and Michael Yarus.
Nearing graduation, Darst applied to medical schools. After two interviews, he realized, “This is not what I want to do.” Unfortunately, the official graduate school application deadline for other programs had passed. So one by one, Darst called chemical engineering department chairs across the country. “I remember some people being really rude: ‘I only take students smart enough to apply on time,’” Darst says.
Then he called Stanford University, whose chair told him, “‘Sounds great. Send an application.’” A week later, Darst was invited to visit. Five years later, he finished his PhD there in biochemical engineering. His thesis wrapped up, Darst used his remaining time in the lab to indulge in “crazy experiments.”
For an electron microscopy experiment, Darst enlisted the help of a graduate school acquaintance, who one day mentioned that his mentor needed another person on an RNA transcription project. Darst told his friend, “Excuse me,” and walked to the office of Roger Kornberg, securing a postdoctoral position studying RNA transcription, the process of making RNA from DNA.
Prokaryote Pincers
To stay alive and perform its duties, a cell must deploy various specific genetic instructions on demand. All organisms deploy some version of RNA polymerase to transcribe the DNA into RNA. The newly made RNA may have a job to do, or it may be translated into working proteins.
Nearly 20 years later, Kornberg would win a Nobel Prize for, among other things, a detailed X-ray crystal structure of the yeast RNA polymerase II caught in the act of transcribing RNA from DNA. That work was just beginning at the start of Darst’s postdoc in 1987
At the time, a three-dimensional (3D) crystal structure of the transcription machinery seemed out of reach, and an X-ray solution, even more challenging. So Kornberg came up with the idea of growing single-layer crystals for electron crystallography. The new technique worked with test proteins, but not for RNA polymerase. Then Darst figured out that the base lipid monolayer could be doped with positively charged lipids to attract a layer of negatively charged RNA polymerase.
In two years, Darst published the first low-resolution structures of E. coli RNA polymerase in combination with the sigma factor molecule that guides it to the correct starting place on the gene, a molecular complex called holoenzyme. It was a big deal.
Scientists knew RNA polymerase had a vague Pac-Man-like shape. The new bacteria structure revealed a more distinctive crab claw shape, with a channel for DNA transcription running between the two pincers. The next year, the team applied the new approach to yeast RNA polymerase II. It took 10 more years of work by others in the Kornberg lab to determine the yeast RNA polymerase II structure at high resolution by X-ray crystallography.
Meanwhile, Darst had met and married Elizabeth Campbell, who was accepted to the microbiology PhD program at The Rockefeller University. They moved to New York in 1992. Darst joined the faculty. Campbell finished her PhD and joined his lab, where she now heads research on transcription in mycobacterium tuberculosis, the bacteria that causes tuberculosis.
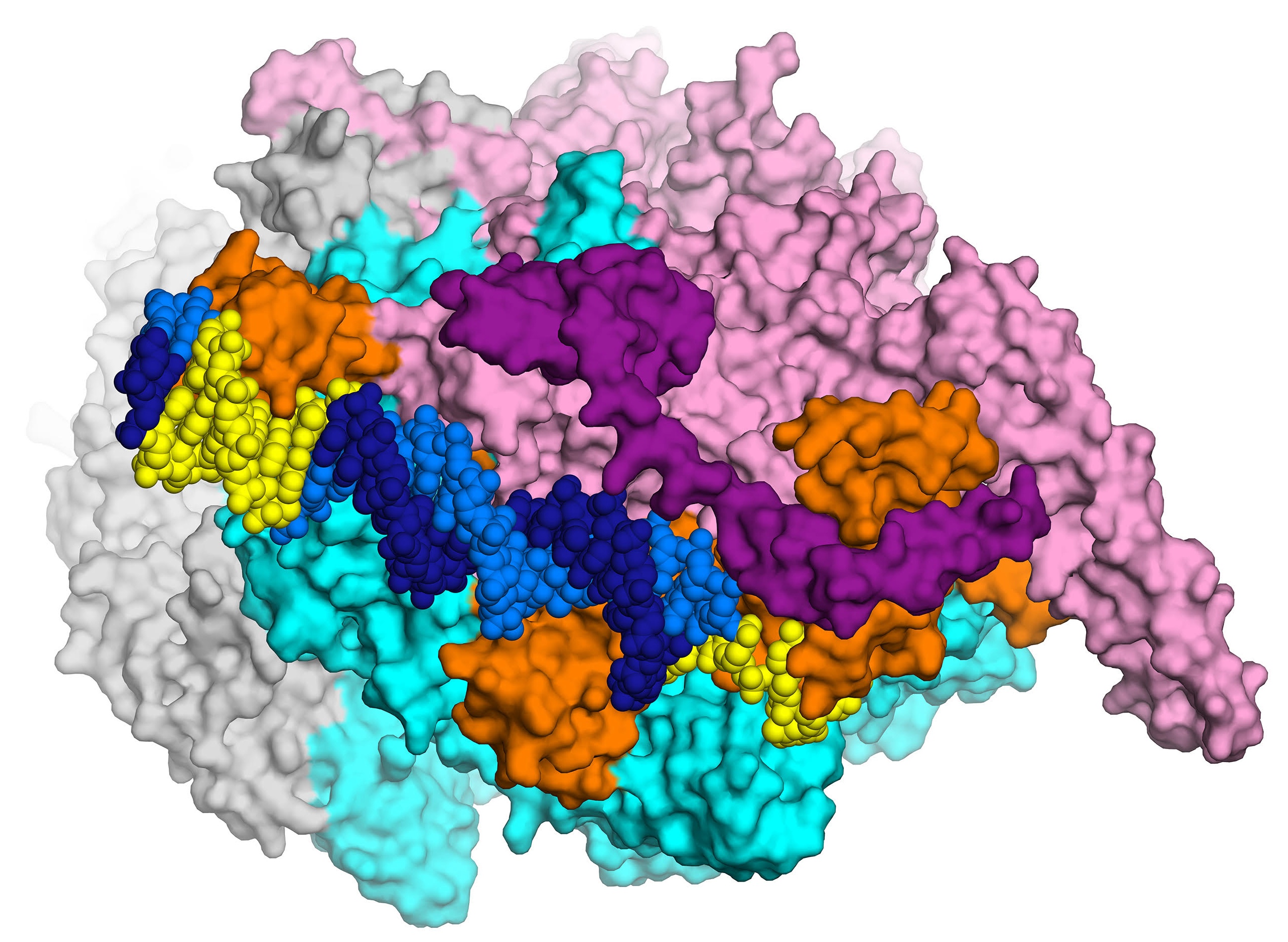
The first high-resolution (2.8 Å) X-ray crystal structure of the transcription initiation complex of Mycobacterium smegmatis will enable structure-based drug design for tuberculosis, thanks to Elizabeth Campbell and her team in the Darst lab. The complex includes RNA polymerase (gray, cyan and pink subunits), the sigma subunit (orange), and promoter DNA (dark and light blue). Courtesy: E. Campbell.
Darst planned to concentrate on electron microscopy (EM) to tease out more details of prokaryotic transcription. Under the influence of the neighboring labs of Stephen Burley and John Kuriyan, Darst transitioned into X-ray crystallography. In 1999, he published the first detailed crystal structure of bacterial RNA polymerase. The 3.3-Ångstrom resolution was detailed enough to make more accurate functional predictions. Darst had abandoned EM about a decade later. As long as they had to do the hard work of growing crystals, he reasoned, they might as well end up with higher resolution results.
As many as one-third of people in the world are infected with tuberculosis (TB), and drug-resistant TB is a growing problem. One first-line treatment, the rifamycin class of antibiotics, targets RNA polymerase. In 2017, Campbell’s group in the Darst lab reported the first detailed structural analysis of RNA polymerase in a close relative of M. tuberculosis using X-ray crystallography. They are using a structure-guided approach to better understand how the drug works, how bacteria become resistant to it, and how to potentially improve the drug’s action.
Darst’s lab is swinging back to EM, thanks to the recent jaw-dropping advances in EM technology showcased by the 2016 Nobel Prize. Not only can the technology enable atomic-scale resolution without arduous rigid crystals, it also can capture subtle shape changes of the molecule at different stages. Now, the Darst lab mostly uses the new EM.
In the past few decades, Darst and others have been able to determine structures of transcription complexes at stable phases of gene transcription by X-ray crystallography. There are, however, many transient and unstable steps in the process that the new EM technologies may help illuminate, including the mysteries of the paused structure. For instance, in one project, Darst’s lab is working towards structures of the paused RNAPs to understand what makes it pause and how the pausing is regulated.
- Carol Cruzan Morton


































































































