SBGrid and Instruct-ERIC establish strategic partnership to strengthen support for structural biology communities across Europe. Together, Instruct-ERIC and SBGrid support European Members through a range of joint activities, combining their strengths to build a more connected, sustainable, and responsive infrastructure for structural biology in the region of Europe.
History
History
-
2025
-
2024
SBGrid Offshoot Comes Into Its Own

The SBGrid spinoff BioGrids matures, as it surpasses SBGrid in software supported, boasting a collection of over 700 software titles for bioinformatics and other biomedical sciences. BioGrids is now a primary platform for software installation and maintenance at Boston Children's Hospital and has expanded to support biotech and pharma industry users. Learn more about BioGrids.
-
2023

Collaboration and CryoEM Training

A colloboration between SBGrid and Meharry Medical College results in a research training program focused on cryoEM data processing workflows with AI components to help mentors develop skills, knowledge, and best practices. This Continuing Education for Structural Biology Mentors program is NIH-funded and seeks to establish long term support for equitable access. Participants return to their home institutions with a set of tools and continuing support to help foster mentorship of laboratory members and students who are pursuing next-generation biomedical careers in structural biology.
-
2020

Community Building During COVID

SBGrid pivots in response to COVID, expanding existing monthly software webinars and transitioning an in-person workshop to a new bi-weekly series of scientific presentations and software tutorials. This series was free and open to structural biologists worldwide, helping to create a forum for the community to connect during months of uncertainty.
-
2018

Tools, infrastructure, and electron microscopy

SBGrid focuses on building new tools to offer the user more efficient and customized software installation options, with a new Capsule environment that wraps all application dependencies and variables together for a faster and more reliable workflow, and a software installation client for all Linux and Mac users that allows the end user the flexibility to manage which software titles to install and when to kick off updates. An electron microscopy specialist also joins the team, to bring bring valuable expertise to this fast-expanding technology.
-
2016

A Year of Workshops

SBGrid hosts 3 workshops. Quo Vadis 2016 - Advanced Crystallography offered students hands-on training in data processing and model building techniques. The NSF-funded workshop on Data and Software Citation brought together scientists from structural biology, quantitative social sciences, linguistics, astrophysics, geography, computer science, and mathematics to develop norms for software and data citation to improve research and resource sharing. Continuing in this vein, the Connecting Journals to Data Repositories workshop put journal editors, publishers, data repository directors, librarians, researchers, and funders in the same room to establish a roadmap for future cooperation.
-
2015

SBGrid Databank

SBGrid launches the SBGrid Databank (SBDB) to preserve and disseminate primary experimental X-ray diffraction datasets that support scientific publications. Fifty member laboratories contributed 117 datasets for the pilot collection described in a Nature Communications publication Nat Commun. 2016. 7:10882. SBGrid is expanding this project in collaboration with Mercè Crosas and Dataverse with support from the Leona M. and Harry B. Helmsley Charitable Trust. Read more
-
2013

eLife and XSEDE

A paper providing a comprehensive description of SBGrid is published in eLife. SBGrid’s Dr. Jason Key becomes a member of the XSEDE Campus Champion program to support the most demanding electron microscopy computations and make high performance computational resources available to the SBGrid community.
-
2012

SBGrid Formalizes Charter

SBGrid rolls out its webinar program featuring software tutorials from a different developer each month. Recordings are publicly available on the YouTube.org/SBGridTV channel. With support from the National Science Foundation SBGrid Policy Fellow, Andrew Morin develops a guide to software licensing (PLoS Compu Biol. 2012;8(7):e1002598) and advocates for better disclosure of source code (Science. 2012 Apr 13;336(6078):159-60.). SBGrid extends software support for pharmaceutical companies with a customized collection of software packages. Membership growth continues at about 28% per year.
-
2010
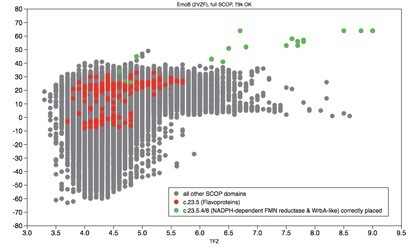
Wide Search Molecular Replacement

SBGrid establishes a Virtual Organization (SBGrid VO) within the Open Science Grid (OSG) and deploys a grid computing portal. Specialized workflows to perform molecular replacement against 100,000 protein domain structures (Wide Search Molecular Replacement) and low resolution DEN refinement are introduced to all members of the structural biology community. SBGrid becomes one of the top OSG users (outside of high-energy physics users) and utilizes ~5,000,000 CPU hours per year. In 2011 SBGrid hosts the Open Science Grid All-Hands Meeting at Harvard Medical School.
-
2008

Quo Vadis Structural Biology?

SBGrid holds its first user meeting: Quo Vadis Structural Biology? (from Latin: where is structural biology heading?). The meeting attracts approximately 300 participants and incorporates a structural biology symposium and three workshops: scientific programming with Python; molecular visualization with Maya; and OSX programming. SBGrid holds subsequent meetings in Boston (2009, 2013, 2014) and, in collaboration with Daniel Panne from Grenoble, organizes the 2010 EMBO Practical Course: Scientific Programming and Data Visualization for Structural Biology, held in Heidelberg, Germany.
-
2006

SBGrid Expands
SBGrid expands to include 37 laboratories located at 14 different institutions: Brigham and Women’s Hospital, Caltech, Cornell, Dana-Farber Cancer Institute, Harvard Medical School, Harvard University, Immune Disease Institute, Rice University, Rockefeller University, Rosalind Franklin University, Stanford, Vanderbilt University, Washington University School of Medicine, and Yale University. SBGrid also establishes itself as an HMS NIH-complaint Service Center. The Center develops an end-user licensing agreement (EULA) in cooperation with the Harvard University Office of Technology Development (OTD) to formalize its relationships with Consortium laboratories.
-
2004

Support For OSX Introduced

In response to demand from Mac users, SBGrid re-compiles the majority of its applications to run under OS X. Today approximately 50% of members use Macintosh computers. The initial collection of OS X application was compiled by Dr. David Gohara.
-
2002

EM & NMR

Sliz and Harrison relocate to Harvard Medical School, several additional research groups join SBGrid, and software support is initiated for electron microscopy (EM), nuclear magnetic resonance (NMR) and other structural biology techniques. SBGrid also begins charging user-fees to recover operational costs.
-
2000

Harvard/Children's Hospital/Yale
SBGrid is established by Dr. Piotr Sliz as a home-grown solution to the challenge of supporting and maintaining a few dozen X-ray crystallography applications that run on SGI IRIX and Linux workstations. The first laboratories supported are those of Stephen Harrison and the late Don Wiley, at Harvard University and Children’s Hospital Boston, and Ya Ha and Karin Reinisch at Yale Medical School. This support includes other existing collaborative software projects in X-ray crystallography, including CCP4 and later PHENIX — both of which continue to contribute to SBGrid.
