The PARP Family Family
John Pascal
Université de Montréal,
Published February 28, 2019
Genetic damage can set the stage for normal cells to turn into cancer. So it may be surprising that blocking a DNA damage repair molecule can kill some cancer cells. John Pascal became intrigued by one of these molecules just as it came on the biomedical radar as a cancer target.
“Inhibiting PARP-1 is exquisitely toxic for cancer cells deficient in certain types of damage repair,” says Pascal, a structural biologist at Université de Montréal in Canada. “Normal cells can largely get by without it.”
Yet, it was unclear how PARP-1 contributes to DNA damage repair in normal or cancer cells. These big unknowns attracted Pascal’s attention. Sweetening the deal, PARP-1 belonged to a larger family of PARP enzymes with mysterious roles in multiple cellular functions. The questions surrounding PARP-1 and its relatives beckoned when Pascal established his first academic laboratory.
“We became a PARP lab,” Pascal says. “We’re PARP people.”
The medical relevancy of PARP enzymes shaped the lab’s structural biology approach. To fight cancer, inhibitor drugs target PARP-1’s catalytic domain, which organizes other proteins around the site of DNA damage to expedite the repair process. The cancer connection motivated the non-profit Structural Genomics Consortium to undertake the catalytic domain structures of PARP family members. Funded by member drug companies and others, the Consortium carries out basic science and makes the data publicly available to accelerate development of new therapies for disease.
So Pascal’s lab took on the other domains that directed PARP-1 on where to go and what to do. “It’s the regulatory domains that dictate what PARPs do in biology,” Pascal says, such as recruiting and localizing PARP-1 to a single-stranded or double-stranded DNA break.
For this difficult problem, they took a divide and conquer approach to study the large floppy molecule, crystallizing individual domains and working their way up to the larger structure. After about six years, they put it all together in a 2012 paper in the journal Science. With all of the essential working parts —the catalytic domain and three regulatory domains bound to a DNA break—it was the biggest structure of PARP-1 at the time.
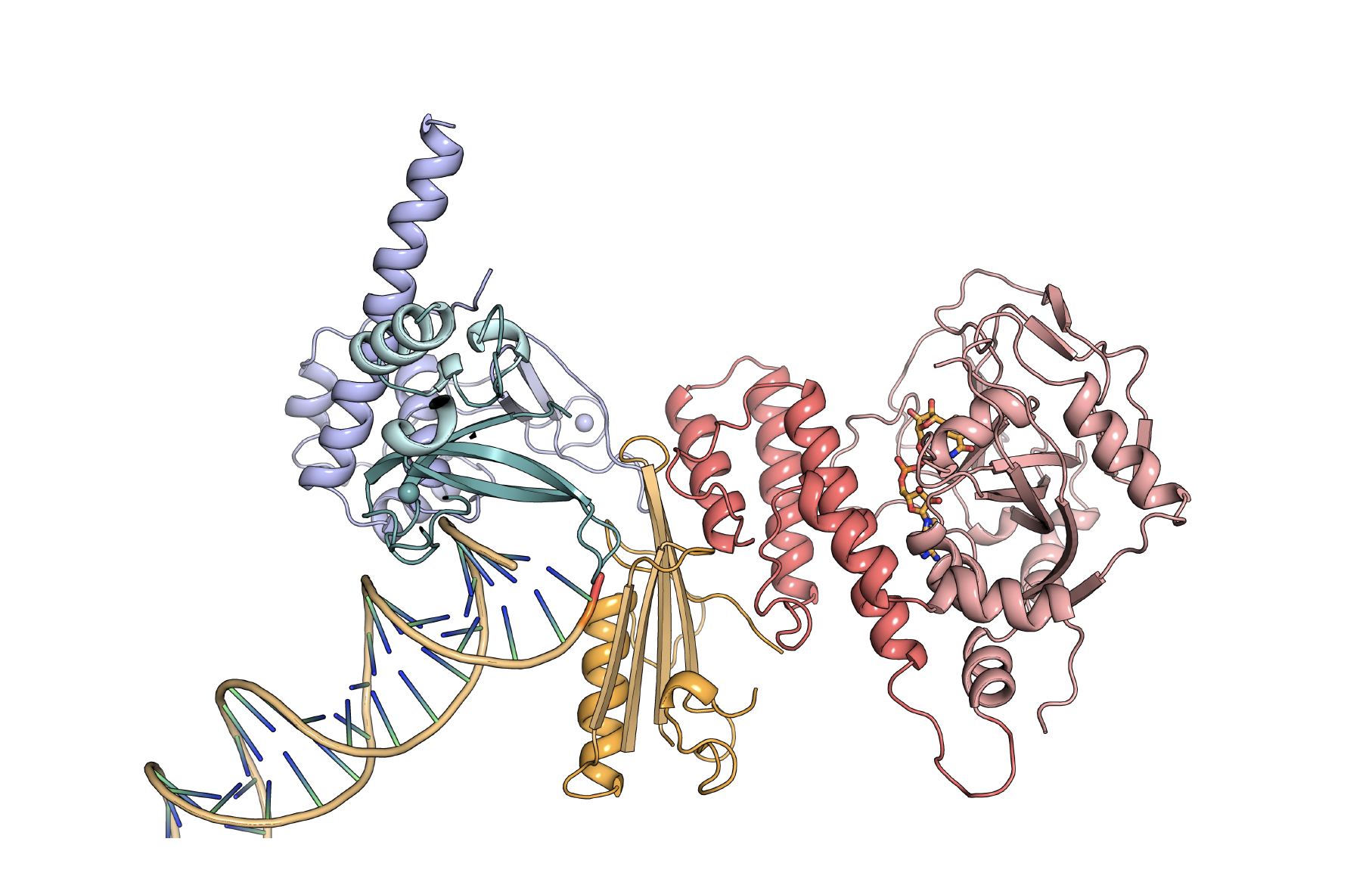
The essential domains of the PARP-1 (Poly (ADP-ribose)- polymerase 1) structure are bound to a DNA double-stranded break. The regulatory DNA damage detection modules (teal, purple, and orange) are connected to the catalytic domain (red) through an allosteric network of interactions. (Image courtesy J.Pascal).
“The PARP-1 project defined who we are and put us on the map,” Pascal says.
In the Science paper, they proposed a model of how PARP-1 works to facilitate repair. The protein folds back on itself when it binds to the site of DNA damage, putting distant regulatory and catalytic domains next to each other. Parts of the molecule directly recognize the DNA damage. Then other parts recognize the DNA-bound parts, and ultimately turn on the catalytic domain so that it can signal the presence of DNA damage and contribute to repair.
When PARP-1 binds and assembles on the DNA damage site, the domain crosstalk induces a change in the fold of one subdomain that flips the switch for 1,000 times more activity. The activated PARP-1 synthesizes large polymers of ADP-ribose that recruit repair factors to the DNA break.
“It’s like a multicomponent signaling pathway all in one polypeptide,” Pascal says. “It’s probably the coolest enzyme I’ve studied.”The study paved the way for designing a new generation of PARP-1 inhibitors that could disrupt the domain-domain interactions required to form an active PARP-1 conformation, wrote other scientists in a commentary accompanying the 2012 Science paper. That’s important in part because some of the cancers treated with PARP-1 inhibitors develop drug-resistant mutations.
The lead researcher on the 2012 Science paper was Marie-France Langelier. A lab can be like a family, but as husband and wife, Pascal and Langelier literally are family.
They met as postdoctoral fellows at Harvard Medical School in Boston. Pascal was studying DNA damage repair in the lab of Tom Ellenberger and Langelier was studying RNA polymerase II transcription in the nearby lab of Steve Buratowski.
They soon became partners in both life and science. Pascal accepted a faculty position at Thomas Jefferson University in Philadelphia. They moved, and she helped start the laboratory.
“At first it was a temporary situation to get the lab going,” Langelier says, “but it worked well.” Langelier considers herself a biochemist and works daily as a bench scientist, while Pascal covers the principle investigator duties.
In 2015, Pascal accepted a faculty job at Université de Montréal,, and they moved the lab there, near where Langelier grew up and earned her PhD. Pascal grew up in Tennessee and earned his PhD in biochemistry at University of Texas at Austin.
The lab has put the PARP-1 structure to use, developing assays to identify inhibitors that disrupt the communication between the domains that gather around the DNA damage site. “If we can stop the protein domains from talking to each other, we can shut down the catalytic activity,” Pascal says. A drug that takes such indirect action away from the active site is known as an allosteric inhibitor.
In another line of inquiry, the lab is trying to understand at the molecular level why some PARP-1 inhibitors are better at trapping PARP-1 on DNA breaks than others. That’s particularly toxic for cells deficient in repair pathways, such as those with risky BRCA mutations, Langelier says.
Pascal is gratified that other scientists have recently begun using structural data from the lab to interpret drug-resistance mutations arising in patients treated with PARP-1 inhibitors. The inhibitors are approved as treatment options for ovarian and breast cancers with BRCA mutations.
Meanwhile, the lab has moved beyond DNA repair and begun exploring other members of the PARP family. In one project, they are exploring PARP-5, also known as tankyrase. It was first discovered as a factor that regulates the length of telomeres, which protect the ends of chromosomes like caps at the end of shoelaces.
To understand tankyrase, the Pascal lab is taking a similar divide-and-conquer approach as for PARP-1, to structurally analyze the regulatory domains, while always building toward the goal of a complete structure. The regulatory domains are entirely different than those found in PARP-1. They include an extended scaffold that recruits other proteins to be modified, and a self-polymerizing unit that regulates catalytic output through an unknown mechanism. The tankyrase catalytic domain is similar to PARP-1 and performs similar actions, such as adding chains of ADP-ribose to itself when activated.
“Many people who I meet are bothered by the need to make a long chain of ADP-ribose,” Pascal says. “Why not use a single ADP-ribose or a more simple phosphorylation modification to get things done? Why the need to create such a large structure? It gets branched and has linear chains like ubiquitin. It’s involved in a number of cellular processes, and an interesting challenge is to understand how the structure of the ADP-ribose polymer can dictate different outcomes.”
The Pascal lab has just begun to explore the PARP family. There are 17 members of the PARP family involved in many biological pathways, ranging from antiviral response to organizing the mitotic spindle. “It’s tempting to view PARP as a curiosity, but I meet people from entirely different areas of biology who find that a PARP enzyme is controlling some key step of a pathway they are studying,” Pascal says. “Then they want to know, ‘OK, what is PARP?’”
-Carol Cruzan Morton


































































































