Rely on This
Sjors Scheres
Medical Research Council Laboratory of Molecular Biology
Published October 26, 2018
But for the luck of a graduate school admission lottery, Sjors Scheres might have followed in his dad’s steps and become a veterinarian. Instead, he headed in a different direction, creating a new generation of software whose importance in biology was footnoted by the Nobel Prize committee in its scientific explainer for the 2017 winners in cryo-electron microscopy (EM).
In The Netherlands, where Scheres grew up, the only veterinary program was oversubscribed. On a whim, Scheres instead chose chemistry and surprised himself by enjoying it. He discovered a fascination with protein structures and soon zeroed in on X-ray crystallography. Halfway through his PhD, he found his ultimate groove when he switched from determining structures in the wet lab to refining the data on computers.
Crystallography seemed too mature for Scheres to base a career. So for his postdoctoral fellowship, he moved his data processing skills to EM in the Madrid, Spain, lab of Jose Maria Carazo. He focused on the development of new image-processing tools.
Among other things, he added a new statistical way of processing EM data, based on “maximum likelihood” methods, for better results to the lab’s original open source XMIPP software package. Scheres was familiar with these methods from his training in methods development for protein crystallography.
In the EM field, these methods were first proposed for single-particle alignment by Frederick Sigworth of Yale University in 1998. The algorithms gave researchers the power to harness more information from EM images. When Scheres added maximum likelihood classification methods to XMIPP, it became the first software program to derive multiple 3D structures from cryo-EM data sets containing mixtures, without needing prior information about what kind of structures were present in the mixture.
Scheres took the next step in software when he moved in 2010 to head his own group at the Medical Research Council Laboratory of Molecular Biology (MRC-LMB) in Cambridge, UK. He called the new package RELION, short for regularized likelihood optimization. It has become one of the most popular tools in EM.

By extending the maximum likelihood methods to a Bayesian formulation, RELION could learn optimal ways of processing the images from the data themselves. This made it easier for scientists to get the best out of their data without being software experts. Scheres has continued to update and expand RELION’s capabilities. In August 2018, his group released version 3. Classifying different structures into homogeneous subsets remains one of the strong points of RELION.
“You can get multiple structures from a single data set, which tells you more about how it works,” Scheres says. The underlying Bayesian statistics are detailed in the January 2012 Journal of Molecular Biology and December 2012 Journal of Structural Biology.
New features in version 3 also allow researchers to squeeze out higher resolution structures from the same set of images, he says. In EM, the force of the electron beam moves the sample, blurring the image slightly. Images are recorded as movies to minimize fuzziness. The new version of RELION corrects any residual blurring a little better than the old version. A forgiving feature added to the new version can rescue some resolution from a not perfectly aligned microscope. The new tools in RELION-3 are described in a Sept. 19, 2018, preprint in BioRxiv.
While developing RELION, Scheres also collaborated with several experimental groups to solve new biological problems in the boom era of new cryo-EM techniques and tools. For example, his group assisted MRC colleague Venki Ramakrishnan with structures of both the yeast and human mitochrondrial ribosomes, reported in 2014 in Science and Cell. In another example, Scheres teamed up with Yigong Shi of Tsinghua University in Beijing, and determined the near-atomic structure of human gamma-secretase complex, which cuts amyloid-precursor protein to form amyloid-beta peptides. The findings were reported in Nature in 2014 and 2015.
Then, in 2016, Scheres switched gears. The single particle field was stabilizing, and the structural biologists with whom he had teamed had become proficient in the new EM techniques and software. The new direction came from a collaboration with neuroscientist Michel Goedert, another group leader at the MRC-LMB.
Goedert had been working for most of his career on tau, the protein that adopts the amyloid filaments that form neurofibrillary tangles in Alzheimer disease. Already in the 1980s, Goedert, together with Tony Crowther, had shown that these filaments could be isolated from human brain and visualized by electron microsocopy. Anthony Fitzpatrick, a postdoctoral fellow in the Scheres group, had a longstanding interest from his PhD studies in amyloids, and was eager to apply the new cryo-EM techniques to tau filaments from Alzheimer brains.
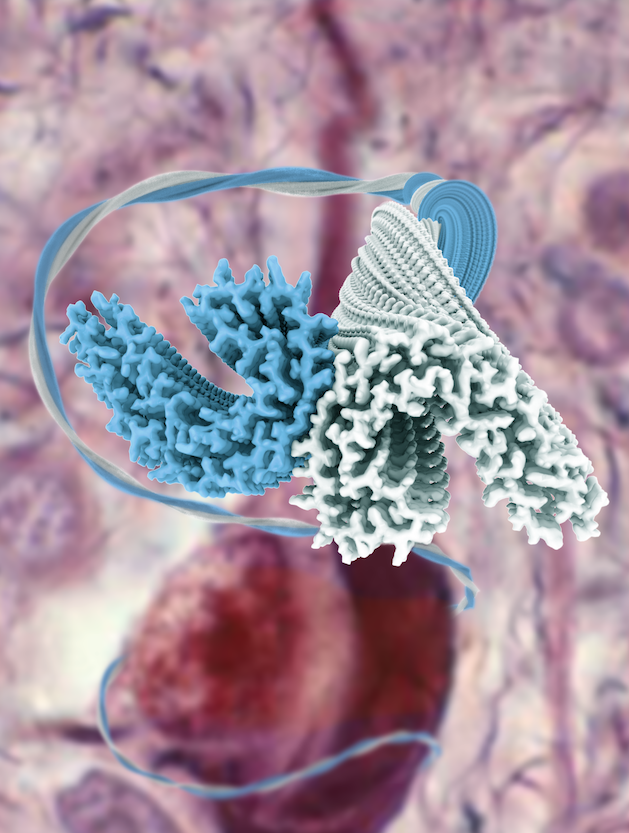
This project depended on the contributions of Shaoda He, a PhD student in the Scheres group, who had expanded the functionality of RELION to deal with helical filaments. Fitzpatrick applied the new methods to tau filaments that were extracted from the brain of an individual with Alzheimer disease, and obtained structures to resolutions that were sufficient for atomic modeling. The findings were published July 17, 2017, in Nature.
The two groups followed up with looking at the tau aggregates from a brain of a person with Pick’s disease. They were testing a hypothesis that misfolded proteins can go wrong in multiple ways, with different folding patterns leading to different diseases.
“We found a completely different (atomic) structure,” Scheres says. “It’s the first time we can confirm the existence of different strain of tau in different diseases.” The study was published August 29, 2018, in Nature.
Now, Scheres runs a split lab, running wet lab experiments on tau in one section and, in the other, making RELION as powerful and user friendly for others seeking to explore amyloids and other proteins themselves.
-Carol Cruzan Morton






























