From Disorder, Function
Julie Forman-Kay
The Hospital for Sick Children, University of Toronto
Published June 29, 2015
After earning a chemistry degree at MIT in 1985, Julie Forman-Kay headed to Yale for graduate work in the lab of structural biology pioneer Fred Richards. Forman-Kay – who says she never grew out of asking “Why?” – had found an ideal mentor. “He was a wonderful scientist who was more interested in questions than techniques,” she says.
The question that stuck with Forman-Kay concerned protein dynamics and disordered states. Though she was initially interested in protein structures, she later realized that many proteins and protein regions function in the disordered state, so over time, her question evolved. Why are some proteins intrinsically disordered, meaning that they naturally — not pathologically — have parts that flop around, noodle-like, rather than snapping into a nice, folded, ordered structure? About 30 percent of the human proteome is intrinsically disordered. “There wouldn’t be so much disorder if it were not functional,” says Forman-Kay.
As a graduate student, she tried to characterize the disordered state structures of fragments of E. coli thioredoxin. She continued this work at the National Institutes of Health in the lab of Angela Gronenborn and Marius Clore, where she finished her PhD, but the research faltered. So Gronenborn gave Forman-Kay a simpler project: solve the structure of human thioredoxin using Nuclear Magnetic Resonance. At the time, around 1990, says Forman-Kay, “it was very sexy to determine structures of proteins by NMR.”
The work helped her land a job in Toronto, where she is now a senior scientist and head in the Molecular Structure and Function Program at the Hospital for Sick Children and professor in the Department of Biochemistry at the University of Toronto. Also, she learned NMR, which is an important tool for studying the dynamics of intrinsically disordered proteins (IDPs). “A lot of biology is in the transitions,” she says. “For me, crystallography freezes out the most interesting aspects of the system.”
The first protein Forman-Kay investigated in her new lab was the drosophila drkN SH3 domain. It turned out to be unstable, in a disordered state about half the time. “I thought, Aha! I can finally do my PhD thesis,” she says.
Since then, Forman-Kay has provided insights into several biological functions that emerge from intrinsic disorder. In studies of the cystic fibrosis transmembrane conductance regulator membrane protein, Forman-Kay found disordered cytoplasmic elements that provide the flexibility needed to integrate various inputs to drive the opening and closing of a membrane channel. Forman-Kay has also found cases where disordered proteins provide precise tuning of cell cycle regulation and protein degradation. “It seems the disordered regions are part of the evolution of complex regulation,” she says. “To think that only folded proteins can provide function is limited. Biology is going to take advantage of all energetic possibilities.”
To support her efforts to understand disordered proteins, Forman-Kay developed a software tool called ENSEMBLE that takes in experimental data, such as NMR data and small-angle X-ray scattering data, and defines a set of conformations that best represent the disordered ensemble. “It isn’t a completely random coil,” says Forman-Kay. “There are structural preferences, but you can’t use standard tools to get one basic conformation, because there is no single conformation that agrees with all the data.”
ENSEMBLE is not yet available as part of SBGrid’s distribution, though that is a goal of Forman-Kay’s. Work is also underway to develop a next generation program.
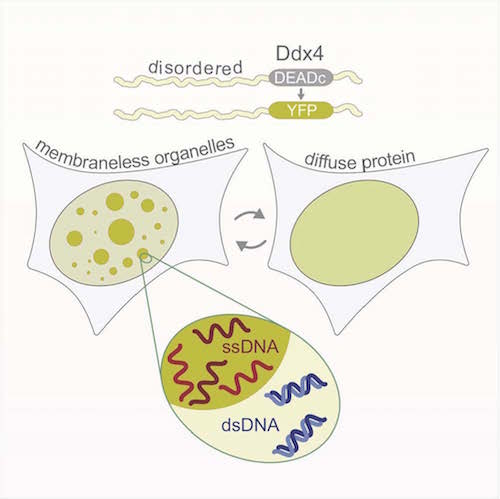
Forman-Kay’s lab is currently focused on understanding the role of intrinsically disordered proteins in sub-cellular structures. In a collaborative 2015 Molecular Cell paper, she describes how a disordered region of the Ddx4 protein forms sub-cellular organelles that are not bound by membranes. The boundaries are formed instead by a liquid-liquid phase separation, a bit like droplets of oil floating in salad dressing. The environment within the droplets is different from without. Functions typically attributed to folded proteins occur inside, such as binding and selectively concentrating single-stranded over double-stranded nucleic acids. “It’s a function that doesn’t require a fold,” she says. “It’s funky but really powerful in terms of understanding cell biology.”
Forman-Kay is continuing work on this model system, working with her husband, University of Toronto biochemist Lewis Kay, on NMR experiments to better understand the interactions that contribute to phase separation. She is also looking at other membrane-less organelles, such as the formation of neuronal granules and nuclear speckles. “Sub-cellular compartmentalization by phase separation completely changes people’s understanding of cell biology,” she says. “I think it’s going to provide a lot of new ways of thinking about biology.”
-- Elizabeth Dougherty































