Cut and Paste
Martin Jinek
University of Zurich
Published January 28, 2015
In January 2013, Martin Jinek published a paper in eLife showing that the CRISPR endonuclease Cas9, molecular scissors that silence the DNA of invading viruses in bacteria, could also be used to edit the DNA in human cells. The work coincided with similar findings from George Church at Harvard Medical School and others, and helped launch the CRISPR gene editing revolution.
The key discovery, however, had come a few months earlier, the result of basic scientific curiosity. Jinek, then a postdoc in the lab of Jennifer Doudna at the University of California at Berkeley, had become interested in understanding the molecular machinery driving CRISPR’s gene silencing. In 2010, he and others in the Doudna lab solved one of the first structures of CRISPR-associated (Cas) proteins bound to CRISPR RNA (crRNA). “In many ways it operated like RNA-interference, but the underlying proteins were very different,” says Jinek, who is now an assistant professor at the University of Zurich in Switzerland. “We were just interested in how this was working at the molecular level.”
Then, in August 2012, Jinek and Doudna provided a detailed description of how a protein called Cas9 works with crRNAs to guide the molecular machinery to the specific sequences of DNA to snip. More importantly, he showed that the Cas9 complex could also be customized. “We showed for the first time that you could program Cas9 to change its sequence specificity and cleave essentially any DNA we wanted,” says Jinek.
From there, CRISPR took on a life of its own, with many researchers focusing their efforts on the development of CRISPR-based genome editing tools. Today, these tools are in wide use not only to develop model organisms with engineered genomes for research, but also in efforts to re-introduce the idea of genetic engineering into human medicine. “It’s amazing how fast this has developed,” says Jinek.
These key discoveries were timely; by 2012, the genome editing field already had traction from efforts to develop earlier technologies such as zinc finger nucleases. “The field was ready for a transformative tool like Cas9,” says Jinek.
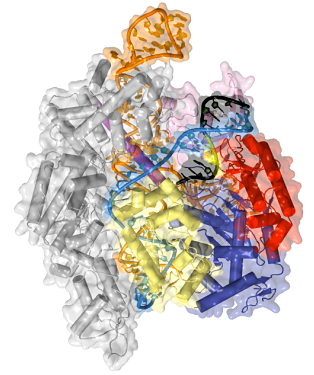
Now in his own lab, Jinek is still focused on CRISPR. He has collaborations with scientists who are applying and developing CRISPR tools. But Jinek’s interests remain in basic science. “Life as we know it is essentially physics and chemistry,” he says. “If we relate this to human health, one change in a gene can decide the life or death of a patient. You can boil it down to interactions between molecules, even atoms. The chemical basis of life still fascinates me and drives my career.”
Jinek’s lab’s primary focus is on understanding Cas9 and its complexes with small RNAs more deeply, using x-ray crystallography as his primary tool. For instance, at first he wanted to better understand how Cas9 uses crRNAs to initially locate a short sequence motif, called PAM, in the DNA to be cleaved. In 2014 he solved the structure of Cas9 in complex with crRNA and a DNA target containing a specific PAM. He found that the PAM provides a gateway for opening up the DNA helix and making the DNA accessible for cleaving. The finding suggests that it may be possible to customize Cas9 proteins to recognize different PAM sequences, adding additional flexibility to CRISPR genome editing tools.
According to Jinek, his studies of Cas9 are just beginning. While the existing tools used for genome editing mostly employ just one Cas9 protein derived from one type of bacterium, the range of Cas9 proteins is wide, of varying sizes and, presumably, functionalities. “There’s a lot of potential for expanding the tool box for genome editing,” says Jinek.
-- Elizabeth Dougherty































