Cosmic Squared
Michael Cianfrocco
University of Michigan Life Sciences Institute
Published June 27, 2020
As one of best selling games of all time, Minecraft and its spinoffs are familiar to hundreds of millions of dedicated fans. But few people know about the video game’s legacy in cryo-electron microscopy.
To be sure, the legacy owes more to the required computing power than the game itself. By the time Michael Cianfrocco was a postdoctoral fellow, cryo-EM tools and techniques had advanced to near-atomic resolution. And the digital imaging data was creating a new technical problem to solve.
Cryo-EM required high-performance computers to convert the images of proteins into three-dimensional reconstructions. “There were all these delays in accessing computer clusters,” Cianfrocco says. “It would take days to wait, and then the job would run super-slow.”
His brother, an engineer, suggested processing the data through Amazon Web Services (AWS). His brother used a tiny slice of AWS for his personal Minecraft gaming, but AWS is vast enough that Netflix also uses its cloud capacity to reach 100 million customers in 190 countries.
The cloud computing option was a conceptual game-changer for Cianfrocco. “You just show up and get as much computing as you want,” he says. “They let you come in with your own software and let you run your job.”
Since then, Cianfrocco has established himself as a scientist with his feet in the lab and his head in the cloud. “I view myself as a structural biologist who builds methods as we need them,” he says. “When I was getting my PhD, everyone had to write their own code. You don’t have to know how to code to do cryo-EM anymore. This lets me keep tinkering.”
Cianfrocco dove into the new coding challenge as a postdoc in the lab. In a 2015 paper in the journal Elife, he and his postdoctoral co-advisor, Andres Leschziner, reported how a cloud computing environment worked with a publicly available yeast ribosome dataset to produce a high-resolution structure. It was labor-intensive, but cost effective. At $50-$1500 per structure, the cloud option made high-performance computing accessible to new labs and could increase the productivity of established labs.
The next step was streamlining the process. Cianfrocco developed a software package that did all the work of submitting data for analysis to AWS and integrated it into the sophisticated RELION data processing pipeline and graphical user interface.
“It’s a neat hybrid,” he says. “It looks like it’s running locally, but it’s actually running on Amazon.” The year Cianfrocco joined the University of Michigan (UMich) faculty, he and his UCSD colleagues reported how their cloud platform worked to solve a 2.2 angstrom structure of ß-galactosidase, a standard sample in cryo-EM. The cloud computing time was more expensive, but not as pricey as buying and installing a similarly powerful computer system. The paper was published in the September 2018 Journal of Structural Biology.
Since being at UMich, Cianfrocco and his UMich faculty colleague Melanie Ohi leveraged Cianfrocco’s experience with AWS to host data processing workshops there. In summers of 2018 and 2019, Cianfrocco and Ohi also brought in participants and guest lecturers to guide scientists with their own cryo-EM data sets using cloud computing resources.
Meanwhile, the next phase of Cianfrocco’s data processing endeavors were sparked half-way through his postdoc. His co-advisors Leschziner and Samara Reck-Peterson moved their labs from Harvard Medical School to University of California, San Diego. There, the groups had a lot of cryo-EM time and were “burning through a lot of structures.”
Fortuitously, UCSD had a large and established supercomputer center, courtesy of the U.S. National Science Foundation. The San Diego Supercomputing Center (SDSC) offered free computing for large computational problems such as cryo-EM. His supercomputing colleagues talked expansively about creating “science gateways,” web sites that allow other scientists to access the computing resource. Even better, supercomputer time was free to scientists who applied for time. NSF funded a gateway demonstration project.
Just before the pandemic hit in March 2020, Cianfrocco was bringing in beta testers to the gateway that he calls COSMIC2 (cosmic-cryoem.org). “The whole goal is to run any software you want,” he says. “All you have to do is hit go. You don’t have to know how to code to do cryo-EM. Let’s give you this platform. You can go deeper and deeper, but you don’t have to if you don’t want to.” Eventually, Cianfrocco plans to use this tool to give anyone who is limited by computing the ability to determine cryo-EM structures.
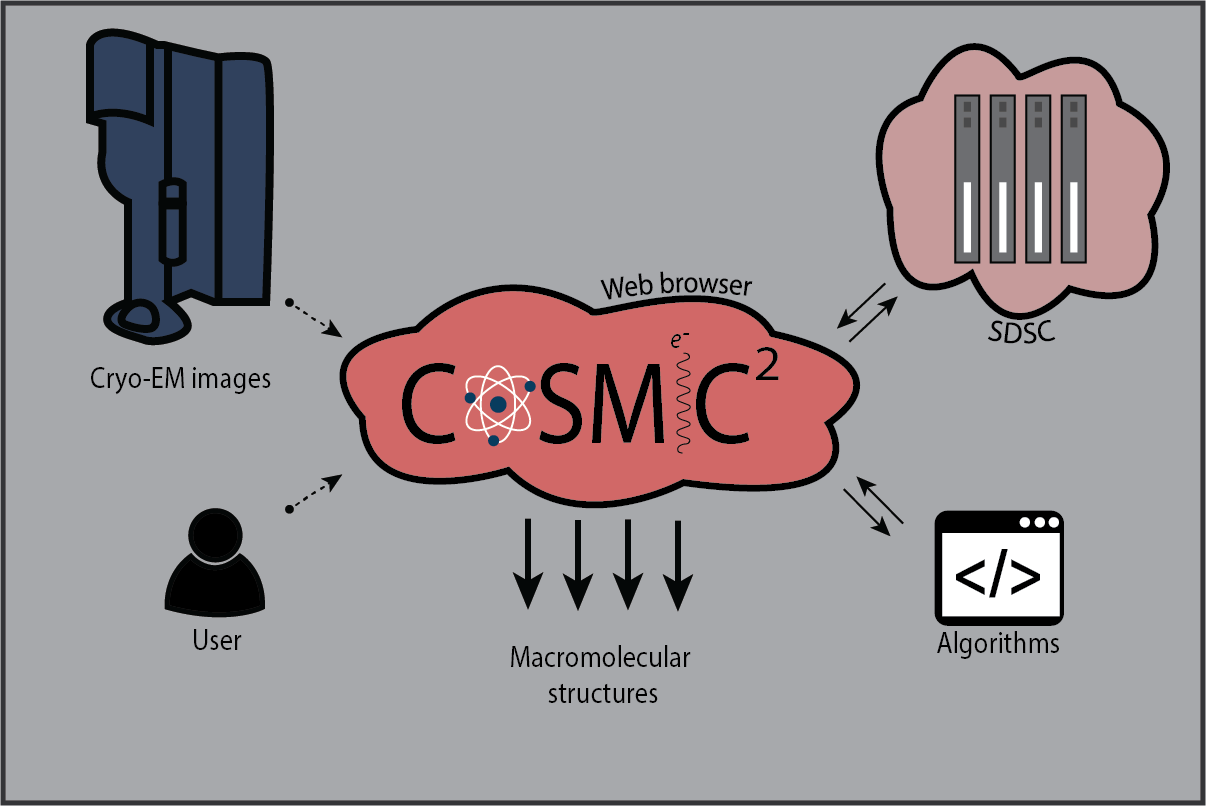
COSMIC2 is a science gateway for cryo-EM. Learn more at cosmic-cryoem.org.
Cryo-EM instruments can collect 10-15,000 images by the time a single project is ready to analyze. With such large amounts of input data, users need to comb through their dataset to remove all bad images, otherwise downstream analysis steps will be negatively affected. “If you introduce a bunch of noisy data into the analysis, it can mess you up later,” Cianfrocco says. “The first steps are important.”
A postdoctoral fellow interested in deep learning joined the lab and led a project to automate the first step in the processing pipeline. Cianfrocco helped train the algorithm by going through about 150,000 images, one micrograph at a time, to teach the machine what’s good and what’s bad. In tribute to the manual assessments, Yilai Li, the postdoc, named the program MicAssess. It is published in the July 2020 Structure.
Now, Cianfrocco says, “you can skip the tedious steps that are hard to learn and require judgement and go straight to the scientific question—‘Is this my protein?’—instead of ‘What is this parameter?’” The software can be downloaded, but he also planned to put it on the SDSC gateway for people to run it on the powerful supercomputer.
For the next step, Cianfrocco wants to bridge the time gap between collecting and analyzing data. “In principle, some deep learning algorithms could run next to the instrument and get results in real time,” he says, with the potential to change the questions scientists can ask the data. He is working with a company that specializes in developing semi-supervised deep learning tools.
“Crystallography is more automated,” he says. “Cryo-EM is very manual. Who knows if this algorithm is the one? Like everything in science, you don’t know which one will stick, but some automatic pipeline will stick in a way that is scientifically rigorous.”

Deep-learning-based single-particle cryo-EM processing pipeline captures human expertise in assessing cryo-EM data. Cover Illustration by Rajani Arora, University of Michigan Life Sciences Institute.
At Providence College in Rhode Island, he majored in biochemistry. He remembers first learning how enzymes work. “When I realized they were doing organic chemistry inside of a protein, it blew my mind,” he says. In his classes, everything made more sense when he could see it. Structural biology was the field where all these pieces came together.
At a summer structural biology program run by NSF, he sampled life as a structural biologist. In a crystallography lab, when he asked about the future of structural biology, a faculty advisor told him about a new technique that in principle could help solve more complex protein structures: Cryo-EM.
At graduate school at University of California, Berkeley, he trained in the lab of Eva Nogales, when cryo-EM was a niche technique that interested few people at the time. Cianfrocco became the last generation of graduate students to use film routinely to capture cryo-EM images for image processing.
The first new digital detectors that helped transform the field were not as good as film, but they could collect a lot more data. His doctoral work focused on transcription biology, which felt like too large and mature field on which to build a new lab. In the Nogales lab, he had listened to five years of talks on microtubules, a topic that occupied the other half of the lab. For his postdoctoral work, he tapped into a little known motor protein called dynein.
Inside cells, motor proteins walk in different directions on microtubule tracks. Cianfrocco wants to understand how motor proteins organize cellular architectures via cargo transport.
“I’m really interested in how they actually move cargo,” he says. Even though motor proteins were discovered a half century ago, “how the molecular mechanics that fuel movement relate to cellular cargo transport, remains poorly defined. It is only within the last few years that the field has begun to reconstitute these complex processes, which will allow detailed structural analysis to relate changes at the atomic scale to cellular and organismal outcomes.”
For example, a long neuronal cell that connects the foot to the spine requires efficient little machines to walk proteins long distance. After all, a protein (or organelle) cannot just use a rideshare application to call for a ride.
For all his cloud-computing skills, Cianfrocco doesn’t play Minecraft. He unwinds by brewing beer, roasting coffee beans, tending extensive backyard garden plots, and cooking with the harvest. These activities have helped to ground him as cryo-EM has quickly moved from low-resolution ‘blob-ology’ to the high-resolution go-to technique the world over.
“Anyone in cryo-EM who recognizes my name would say he’s the cloud computing guy,” Cianfrocco says. “That’s my cryo-EM brand.”
-Carol Cruzan Morton































