Right place, right time
Ernesto Fuentes
University of Iowa College of Medicine
Published January 29, 2021
As a kid, Ernesto Fuentes lived in Brazil just long enough to stock up on the soccer skills he now shares with his kids. His affinity for structural biology may have arisen at the same time, shaped by family trips to see churches and other architectural highlights in nearby countries. Little did he know at the time, he would grow up to bring fresh insights to common building blocks responsible for a dazzling diversity of normal molecular function in living cells.
Back in the United States, Fuentes attended college in Ohio. He intended to become a chemical engineer, like his dad. Then he learned that proteins, like buildings, have distinctive architecture. He pivoted to a biology masters degree and followed up with a PhD specializing in nuclear magnetic resonance (NMR).
His doctoral studies immersed him in NMR and dynamics in more ways than one. His mentor’s lab moved twice, from University of Illinois to State University of New York Buffalo and then to University of Pennsylvania.
As a postdoctoral fellow in a University of North Carolina cancer laboratory, Fuentes established a paradigm for his future lab: Collaborate with colleagues doing the biology.
That leaves Fuentes free to explore fundamental questions about how a common part of many proteins can be involved in so many specialized tasks inside cells.
His postdoctoral work introduced Fuentes to PDZ domains, a small protein region found in more than 250 proteins that cover a broad swath of biology. PDZ domains connect partner proteins to each other in large signaling structures inside a cell next to the membrane in an arrangement that facilitates specific biological action.
PDZ domains somehow put together the protein scaffolding “in the right place at the right time” to get things done, Fuentes says.
Thirty years ago, scientists figured out the basic function: A PDZ domain in one protein grabs another protein by its tail. The tail, or C-terminus, fits neatly into the main PDZ groove. But there are subtleties in that maneuver that dictate exactly what other proteins can connect and what they can do as a result, Fuentes and others are discovering as they develop new ways to learn the rules of the PDZ family specificity.
Since Fuentes joined the University of Iowa College of Medicine faculty in 2006, his lab has been probing the specialized interactions of PDZ domains.
In their research, the Fuentes lab applies multiple integrated structural biology techniques for deeper insight, including NMR, X-ray scattering, crystallography, and cryo-electron microscopy, leaning on a full repertoire of SBGrid software.
“We don’t do biology, but we’re really interested in biology,” Fuentes says. “We’re developing tools that allow us to interrogate biology. We can do that through structure.”
The PDZ domain is found in many proteins and a protein can have one or many PDZ domains. Not only is PDZ a popular domain in proteins, it is a veritable social butterfly. PDZ domains bind to their counterparts in other proteins and mix it up with other ligands. Learning how finely tuned PDZ domains meet up with binding partners will illuminate normal biological signaling and may help develop more precise therapeutics.
PDZ domains help build protein complexes that facilitate different types of cell adhesion and signaling. In particular, they allow epithelial and neuronal cells to form cell-cell adhesion complexes that are critical for normal tissue and neuronal function, Fuentes says. Mutations that affect PDZ activity can have health consequences, including cancer, deafness, neuropsychiatric conditions, and cystic fibrosis.
In a central question, the lab has been asking how selective are PDZ domains in their binding partners, also known as specificity.
To sum up a dozen years of work with three PDZ family members (CASK, TIAM1/2, and Scribble), Fuentes and his colleagues have found that the PDZ groove for a protein tail is just a starting point. Some of the specificity comes from dynamics in the other PDZ nooks and crannies known as subpockets. Not only can certain amino acids wiggle into place better than others on a PDZ domain, the PDZ subpockets will adjust themselves to accommodate a preferred protein partner.
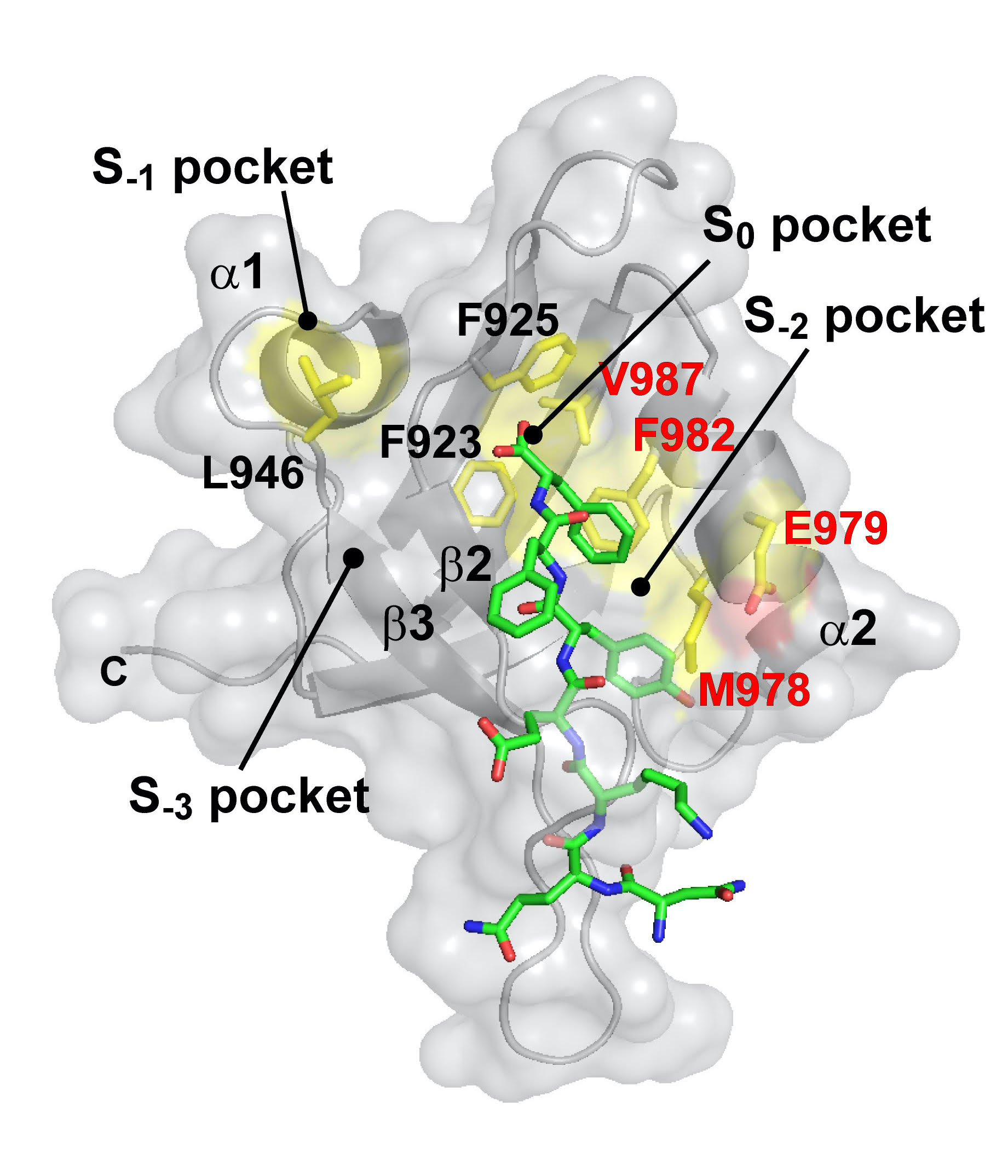
“Changes in the dynamics might be key to understanding specificity,” Fuentes says. For example, a protein with more dynamic range may be able to accommodate more partners and participate in different types of signaling. A less dynamic protein may have a more specific role. That means a mutation could allow more or less dynamic changes, influencing the biological system.
The researchers have experimented with mutations in different PDZ subpockets to observe the impact on binding. Some mutations sparked increased dynamics, also called hot spots, and those PDZ domains favored different or more binding partners.
“The really cool thing, this gave us insight into how evolution can change the specificity of a single PDZ domain,” Fuentes says. “Over time, mutations may be able to broaden or direct the domain to a new set of targets.”
In another twist, they found that some PDZ domains can bind internal motifs and not just the canonical C-terminal tail. “What’s interesting is that it opens up another set of interactions and set of proteins,” he says. “There are not that many examples, and the specificities are not well understood.”
Fuentes found a rare example in a collaboration with a cancer researcher who is studying how epithelial cells normally are held together so tightly. Most cancers originate in epithelial cells, and cancers have to overcome the cell-to-cell adhesion system to grow and spread.
In the project, Fuentes' team was tasked with dissecting an interaction between the proteins SGEF and Scribble involving a PDZ domain. They found an internal PDZ motif in SGEF that binds to one of Scribble’s PDZ domains.
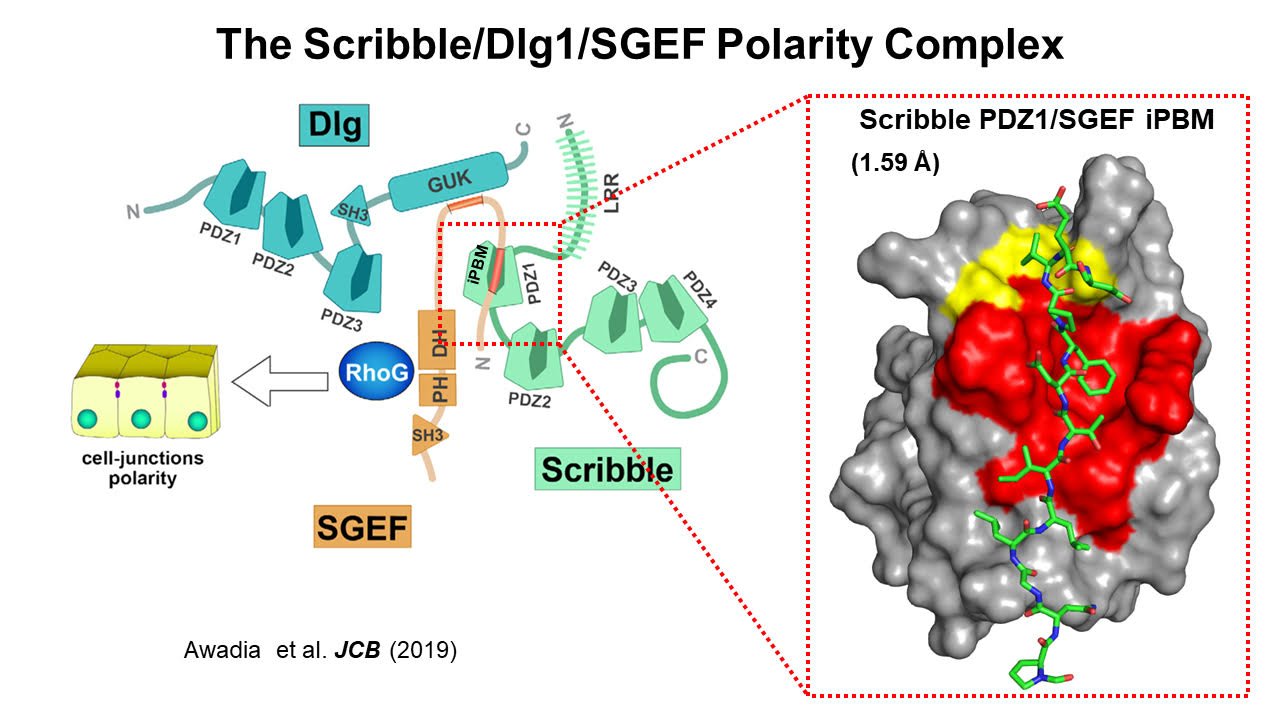
But a crystal structure revealed a new mechanism of PDZ specificity. The SGEF internal motif binds tightly because additional amino acids can latch on to an extra compatible section of Scribble’s PDF domain. The paper was published June 2019 in the Journal of Cell Biology with co-author Rafael Garcia-Mata at the University of Toledo.
The Fuentes lab will work toward a structure of the larger molecular complex with electron microscopy. But the experimental model also opens up a new way to study PDF specificity in more detail using synthetic biology methodologies in collaboration with a research team at Johns Hopkins University.
“We’re now looking at artificial designs of PDZ with the idea of obtaining a deeper understanding of the rules for how nature encodes specificity in a small domain,” Fuentes says. Preliminary work has produced a template protein domain with structural and biophysical properties of a PDZ. “We have a blank slate to design and test the features of specificity,” he says.
In other non-PDZ work from the Fuentes lab, they and their collaborators discovered details of the SrrB two-component signaling system that helps Staphylococcus aureus sense a change from an aerobic to an anaerobic environment.
S. aureus is a common bacterium found in people’s noses and on skin, but inside the body it can turn into a potentially deadly infection. Fuentes struck up a collaboration with a university colleague, Pat Schlievert, who discovered a two-component signaling system important for environmental sensing that allows S. aureus to make the dangerous adaptation to a low oxygen environment.
As they reported in a 2019 paper in PNAS, the Fuentes team found redox active cysteines in a sensor histidine kinase had a big influence on S. aureus pathogenicity.
If there is a theme running through the Fuentes lab, it’s to build an integrative structural biology approach using the tools that best suit the needs of the problem. And to keep an eye on the bigger picture – the biology.
“Structural biology is a part of many studies,” Fuentes says. “It’s not always the central feature. It’s an added method that allows deeper insight into the whole system.”
-Carol Cruzan Morton































