Proteins out of bounds
Gerhard Wagner
Harvard Medical School
Published September 27, 2018
As a physics undergraduate student in Munich, Gerhard Wagner worked on an esoteric atomic measurement of iron in a protein molecule. Then he heard from his supervisor, who was on sabbatical at Bell Labs in New Jersey. There, the same molecule, hemoglobin, the iron-rich protein that carries oxygen in red blood cells, was being probed by nuclear magnetic resonance (NMR) spectroscopy.
The technology caught Wagner’s attention. Why measure a single parameter, as he was doing, when you could measure many aspects at once with NMR and learn so much more, he asked himself. Little did he know, but that thought launched a lifetime of NMR research.
Wagner hadn’t planned on a career in academia. He was born in Czechoslovakia just as World War II ended. His German-speaking family became refugees, fleeing to nearby Bavaria and starting again with nothing. In science, Wagner also helped develop a new field from scratch.
When he graduated from Technical University in 1972, he found a sweet spot in Switzerland—home of his girlfriend, skiing, and a PhD training position with Kurt Wüthrich, one of the early scientists to use NMR on proteins and who later won a Nobel Prize. He spent most of the next 14 years in Zurich.
At ETH Zurich, Wagner launched a family and a career in NMR that soon brought him to the attention of the world’s top structural biologists. At the time, protein structures could only be determined by X-ray crystallography. Wagner helped develop suites of techniques for the fledgling NMR analyses of proteins that made it possible to assign resonances and solve their structures.
Early on, he observed one of the NMR advantages: protein dynamics. “I found a lot of mobility in proteins,” he says. These days, NMR is well known for its unique ability to reveal thermodynamic and kinetic aspects of proteins. More than 40 years ago, those protein wiggles were nearly impossible for some to imagine.
Wagner was testing the new techniques on a cow protein with stable ring-shaped parts (called aromatic side chains in chemistry parlance). Analyzed by NMR, the rings flipped when the temperature went up or down, opening the protein structures by several Ångströms. The unexpected results were controversial.
Wüthrich planned to talk about the new data at a physics conference in Germany. Behind the stage, conference organizer and Nobel Laureate Max Perutz didn’t believe the results and objected to the presentation. Perutz famously had solved the structure of hemoglobin, which also sported aromatic side chains. Another prominent scientist, Robert Huber, who later also was awarded a Nobel Prize convinced Perutz to let the talk proceed as planned, Wagner says. (Later, the same data provided a test bed for the development of molecular dynamic simulations by Martin Karplus.)
After a one-year postdoctoral position at the Massachusetts Institute of Technology in Cambridge, Wagner continued his work in Wüthrich’s lab in a postgraduate teaching and research position called privatdozent. Soon it came time to present Wagner’s latest work, the NMR structure of cadmium-bound metallothionein, a small protein that binds zinc or other metals in plants and animals. Wagner got a worried call in the middle of the night from for Wüthrich. It seems the NMR structure was very different from a crystal structure of the same molecule being presented at the same meeting. Wagner and his coworkers had applied a new technique, proton cadmium correlations, to determine the topology and structure. He scrambled to review the data. It all checked out correct.
“It was also an early time in crystallography, so people could still make mistakes,” Wagner says. “It gave me some visibility. It showed NMR could do a structure and get it right.” Later, the two NMR and crystal teams published a joint paper showing agreement between their final metallothionein structures.
NMR eventually became an accepted method for solving protein structures, especially of smaller proteins in lower concentrations. Wagner took a faculty position at University of Michigan in 1987. There he just had time to develop the first triple resonance experiments, a strategy that is now the basis for most protein NMR, before he was recruited to Harvard Medical School in Boston in 1990.
“When I came to Harvard 25 years ago, I came as an expert in NMR,” Wagner says. “I thought I should also have an important biological project.” He looked for areas with little research and landed on translation initiations, or how a transcribed gene is turned into a protein.
After a gene is transcribed into RNA, a large protein complex grabs hold of the messenger RNA tail to prepare to make a protein. In a 2003 Cell paper, Wagner’s lab reported the structure of the first two proteins in this complex (eIF4E and eIF4G) and how they enable the ribosome to bind to the 5’ end of mRNA and start making protein.
“Then the idea came that oncoproteins have long 5’ UTRs,” or untranslated tails, Wagner said. “Our hypothesis is that nature developed long tails to make it more difficult to translate dangerous proteins.” Other labs have shown that the long tail effectively down regulates some proteins. In cancer, such controls on the oncoproteins may be broken, he says. Restoring them may be a new way to treat cancer.
Wagner and his collaborators screened for small molecules to inhibit some of the translation initiation proteins his lab had characterized. Now they have a major program to improve the compounds they found with anti-cancer activity.
These days, NMR remains well suited for characterizing smaller proteins and to catch the gymnastic moves of molecules in their active biological roles. In particular, Wagner believes NMR has potential to illuminate RNA in complex with other proteins. “Another area where NMR may have some impact is proteins that are only partially folded,” he says.
For example, the nuclear factor of resting T cells has a long unstructured tail hanging in the cytoplasm of cells. The tail has many phosphorylation sites, keeping it in a resting state. Working with another DFCI collaborator, Wagner’s group developed a new NMR technique (known as direct 15N detection) to elucidate a key internal step in activating T cells, which is important to fight infections but also needs to be regulated to prevent rejections of organ transplants. In a collaboration with Haribabu Arthanari at Dana-Farber Cancer Institute, the team identified a key molecular interaction in activated T cells. The Wagner lab is developing small molecules that inhibit the interaction, ideally targeting a protein-protein interaction with fewer side effects than current anti-rejection drugs.
In another project, Wagner’s group has found a better way to study membrane proteins in a more natural environment. A team led by postdoctoral fellow Mahmoud Nasr improved the design of tiny nanodiscs, a popular model of cell membranes used to study proteins entering cells.
The Wagner lab’s modification of the design made the nanodiscs more stable using some fancy chemistry bonding membrane scaffolding proteins. The resulting covalently circularized nanodiscs can be precisely sized from 9 to 50 nanometers and used to study viruses and other membrane interactions by adding a receptor. In their first published demonstration in 2017 in Nature Methods, the team and its collaborators observed polioviruses opening a putative pore and injecting genetic material through the nanodiscs, as the virus might do when it infects a cell.
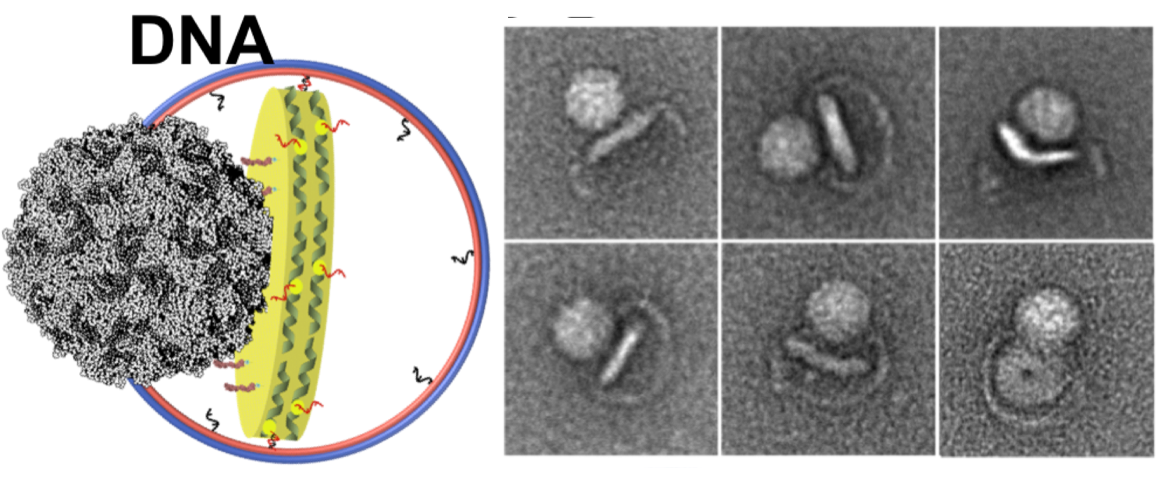
Left: A cartoon illustrates the poliovirus (ball) attaching to its receptor CD155 in the membrane interior of a DNA-corralled nanodisc. Right: The EM micrograph series shows the sequence of the poliovirus engaging with the membrane, ejecting RNA through the nanodisc. Then the empty virus leaves behind a putative ejection pore.
When Wagner moved on to study HIV and the process by which it fuses with a cell membrane, the nanodiscs became too big for NMR. The ongoing work requires cryo-EM analysis. Collaborating with William Shih’s lab at the Wyss Institute, Wagner’s group developed a larger nanodisc design with the lipid bilayer inside DNA scaffolding. He sees many possibilities ahead with the new technology, including nanodisc complexes.
-Carol Cruzan Morton































