Data Whisperer
Karolin Luger
University of Colorado Boulder
Published May 30, 2018
Like others in her sporty college town, Karolin Luger heeds the call of the mountains she can see from her laboratory at University of Colorado Boulder. Trail running and a daily 20-mile round-trip bike commute at altitude have kept her fit, but she would like to put to rest the rumors that she runs her research collaborators ragged on strenuous hikes. “All these crazy stories,” Luger says. “I only did that once.” And lately, she says, she’s the one who has a hard time keeping up on group outings.
Her collaborators can relax, but Luger wants to see more dynamic motion in the chromosome structures she studies. After all, they have to move to do what they do. Luger’s research addresses a fundamental question: How does the human genome store information and then access that information at the appropriate time? Part of the answer, she and her collaborators have learned, goes back billions of years.
In animals, a full genome must be squeezed into the nucleus of each cell. In humans, that means six feet of DNA must fit into something less than one-fifth the size of what the human eye can see. This can happen thanks to the compact packaging of DNA into an assembly called chromatin. Chromatin consists of a long strand of tightly twisted DNA wrapped twice around repeating flat spools of histone proteins, like a string of beads. The protein-DNA spools, or nucleosomes, are the basic repeating unit of chromatin. They are further folded and arranged into a higher order structure that Luger’s lab also studies.
“Imagine a super-long sewing thread and wrap it around a million tiny hockey pucks,” Luger says. “It’s actually really a miracle that there are not knots and tangles.”
Chromatin controls access to genes in ways Luger and her colleagues are still learning. The tightly packed hockey pucks, or nucleosomes, must unspool to activate certain working genes, to duplicate DNA during cell division, or to repair damaged DNA, and then rewrap. “We’re still far away from figuring out how that works,” she says.
Complicating matters, genes can be independently activated or silenced by small chemicals that tag the histone spools or the DNA itself. Luger’s lab is trying to figure out how those small epigenetic tags affect the packaging of the hockey pucks, especially during development, as the same genetic blueprint produces such different cells—brain, heart, muscle, liver and skin, blood, bone.
Luger grew up Austria in a landscape dominated by mountains and science. Her father was an engineer, and her brothers were interested in electronics and physics. Luger was first drawn to botany and zoology. She studied microbiology at University of Innsbruck. Bleak job prospects and a rewarding undergraduate experience in a biochemistry lab inspired her to pursue a PhD in protein engineering at University of Basel, Switzerland.
Given a free hand by her mentor, she wanted to learn if a protein could fold normally if it came out of the protein-translating ribosome backwards, somewhat like a breech birth. (The answer is yes, it can.) She developed a new technique to test the idea, joining two ends of a gene together and cutting it open in a different place. The results were published in Science in 1989. Her studies with such proteins ended with her doctorate, but others have used the information in evolutionary studies to detect genes that make proteins of similar structure and function, but whose gene sequence might be permuted in this manner.
Her PhD program had required her to take classes in structural biology. To satisfy a new craving to learn crystallography, she joined the lab of Timothy Richmond at Swiss Federal Institute of Technology as a postdoctoral fellow. Her project resulted in the long-sought high-resolution structure of a nucleosome at 2.8 angstrom, published in Nature in 1997.
“I had no idea what a super-gnarly problem it was,” she says. “I learned the hard way, but it was a good learning experience.” She also learned that she had a visual brain highly suited to structural biology and developed the methodology (now used in many labs world wide) to make nucleosomes for crystallization and for biochemical and biophysical studies.

She joined the Colorado State University (CSU) faculty in 1999 and became a Howard Hughes Medical Institute investigator six years later. In 2015, she moved her lab to the University of Colorado, Boulder. Luger continues to collaborate with CSU faculty at the Institute for Genome Architecture and Function, a research consortium she and two fellow CSU women faculty members founded in 2015.
Her lab’s work has broadened from solving crystal structures to using cryo-EM, and also to asking scientific questions using structural biology as one of many tools, including live cell experiments. Luger likes to find new ways to see something that wasn’t previously observable. “I almost like the process of doing science more than the actual results,” she says. “I love the process of discovery, to figure out mechanisms and trick the system to tell me its secrets.”
She has also become more action-oriented. “We can stare endlessly at structures,” says Luger, who admits she still is blown away by the transformative power of hydrogen bonds and amino acid packing. “But we need to think about function even more. Most of the structures we study are machines and have to move. We are more and more interested in catching these machines in the act of going about their business. It’s immeasurably harder than looking at the static structure, which is already hard enough.”
Luger describes the narrative arc of her lab’s research path as a “directed random walk.” She adds, “My feelers are open all the time to see what I can learn from others. I just love collaborating with people and integrating ideas out of left field into my research program. It drives people in my lab crazy sometimes, because they are the ones that have to do the hard work.”
In one collaboration reported in Science in 2006, Luger and her colleagues examined a crucial binding point between a protein on the virus that causes Kaposi’s sarcoma and a pair of histone proteins in the nucleosome. “This was the first time it was shown how nucleosomes can serve as docking stations for other proteins,” Luger says. “As far as the virus goes, this gives its genome a nifty way to hitchhike on the host’s chromosomes to escape its defense mechanisms.”
Two years later, Luger and her collaborators used new techniques to mimic the chemical marks on tightly wound DNA-protein complexes that silence gene expression. They reported that repressive epigenetic marks on histone proteins cause nucleosome arrays to become more compacted, while activating methylation did little to the chromatin. The findings were published in Nature Structural & Molecular Biology.
Scientists have wondered about the origins of nucleosomes. Luger and her collaborators looked for answers in archaea, an extended family of ubiquitous single cell organisms thought to be the ancient precursors of eukarya. In this study, they determined the structure of nucleosomes of a species thought to be endemic to Icelandic hot springs.
“One surprising thing we have learned from our studies of archaeal chromatin is that the way DNA is bent into shape by histones is older than eukaryotes,” Luger says. “This principle is being used by an ancient domain of life, the archaea. But unlike in eukaryotes, were we have defined particles, archaea have kind of 'continuous nucleosomes' that look like a slinky. What really surprised me was how similar the organizing principle is between these two domains of life that are separated by billions of years.” The findings were reported in Science in 2017.
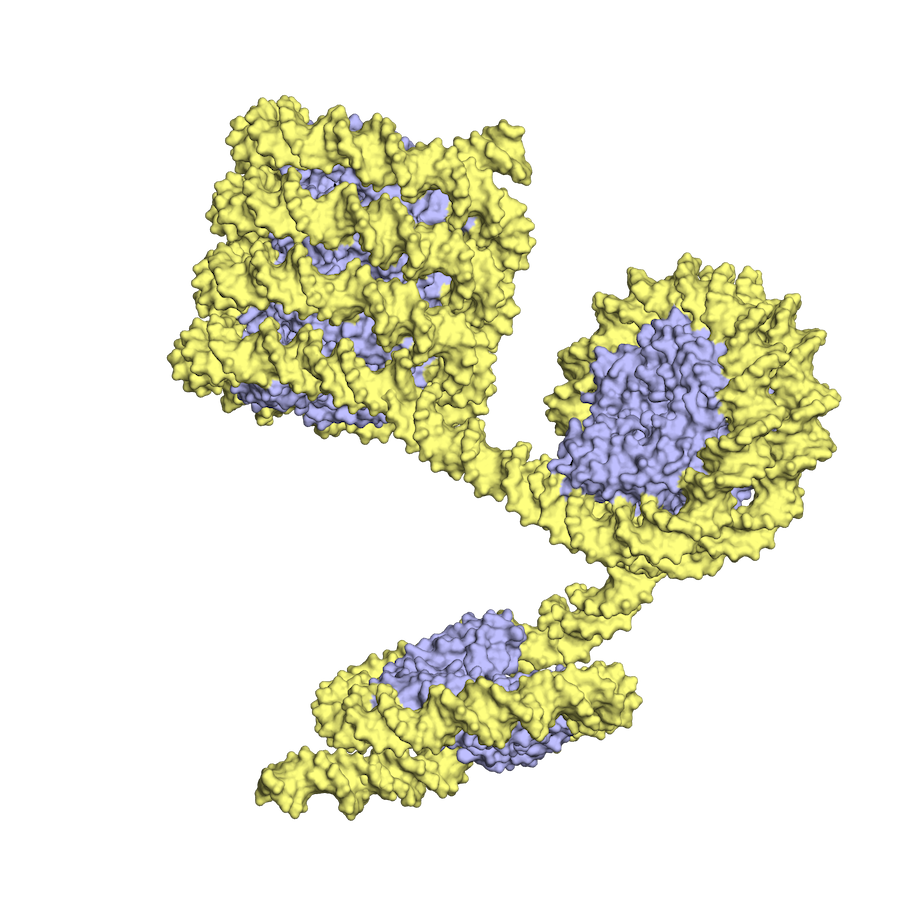
In archaea, the histone-DNA complex can vary in the length of wrapped DNA, as shown here, while eukaryotic nucleosomes have about 2 DNA wraps each. (Images courtesy of K.Luger and CU Boulder)
Other projects include looking at how a dividing cell makes and assembles new nucleosomes for the newly replicated genome. The Luger lab is also interested in DNA repair proteins targeted by promising new anticancer drugs, seeking to learn how proteins know the DNA is damaged and then how the proteins help repair the damage. One project aims to develop an “outlandish tool” to target nucleosomes for gene editing. “It’s always fun to have at least one ‘crazy’ project,” she says. When taking on new lab members, Luger looks for new perspectives, such as enzymologists, “hard core physicists,” or cell biologists, who look at things differently. It’s one way of fighting what she considers the most dangerous threat to science in any lab: Confirmation bias, or the innate human tendency to interpret results in a way that confirms one's hypotheses, especially when results contradict each other are unexpectedly confusing. “I find myself all the time saying let’s assume the whole premise is wrong and let’s rearrange our assumptions,” Luger says. “I don’t believe results lie. It’s literally like a puzzle. A couple of pieces kind of look like they should fit but they don’t, and we have to start over again.” “That’s my mantra,” Luger says. “Problems are three-dimensional. You have to walk around and look at them from another viewpoint. You have to constantly check your premise. Even if nothing fits, people (including me) are reluctant to let it go. I keep telling my coworkers: Your results are telling you something. You have to listen. If they are confusing, it’s not the results’ fault. I really do believe the systems we’re studying are trying to communicate with us: ‘I’m right here. This is what I’m doing.’ I feel their desperation, which is mutual. Most people in lab are quite sick of hearing me say this, I am quite certain.”-Carol Cruzan Morton































