AlphaFold protein structures & ChimeraX cryoEM modeling
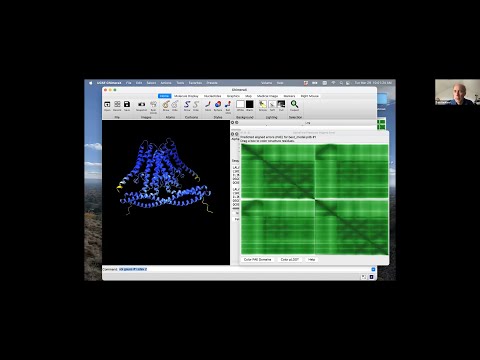
Presenter: Tom Goddard, UCSF Resource for Biocomputing, Visualization, and Informatics - Topic: Using AlphaFold protein structures in ChimeraX for cryoEM modeling
Linked to presented materials: https://www.rbvi.ucsf.edu/chimerax/data/sbgrid-mar2022/alphafold_pae.html
Learn more on the ChimeraX website:
https://www.rbvi.ucsf.edu/chimerax/data/alphafold-nov2021/af_sbgrid.html
This talk was presented as part of the SBGrid Spring Mini-series - Cryo-electron microscopy of membrane proteins: from sample to structure. See the full series lineup at https://sbgrid.org/webinars/
Organized by
Prof. Jamaine Davis, Meharry Medical College
Prof. Piotr Sliz, Harvard Medical School
Prof. Patrick Sexton, ARC Centre for Cryo-electron Microscopy of Membrane Proteins, Monash University
Recorded on March 29, 2022
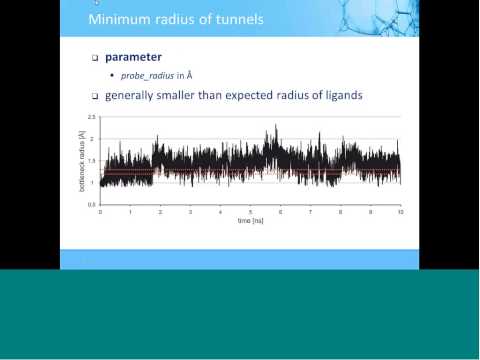
Topic: CAVER 3.0: Analysis of Tunnels in Static and Dynamic Protein Structures
Presenter: Jan Brezovsky, Ph.D., Team Leader, Loschmidt Laboratories, Czech Republic
Host: Jason Key
Recorded on June 13, 2013
CCP4 Molecular Graphics (CCP4-MG)
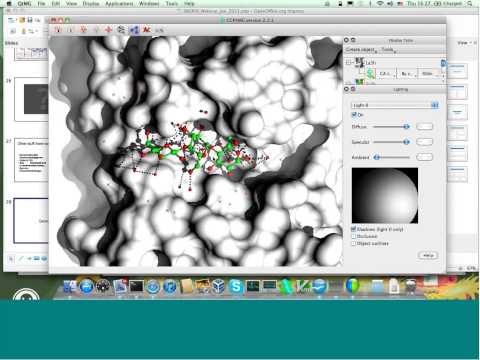
Topic: The CCP4 Molecular Graphics Program
Presenter: Stuart McNicholas, Research Fellow and CCP4-funded Developer, Dept of Chemistry, University of York
Host: Piotr Sliz
Recorded on January 24, 2013
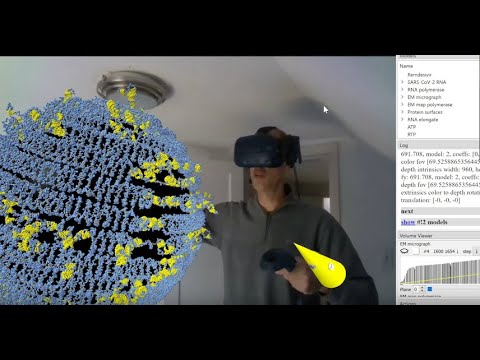
Topic: Making Augmented Reality Videos with ChimeraX
Presenter: Tom Goddard, University of California, San Francisco, Resource for Biocomputing, Visualization, and Informatics
The Zoom recording of this presentation is pretty choppy. We recommend you also check out the video Tom recorded and posted on his own channel here: https://youtu.be/dKNbRRRFhqY
Slides from talk available at https://www.rbvi.ucsf.edu/chimerax/data/sbgrid-july2020/
Host: Pete Meyer
Recorded on July 7, 2020
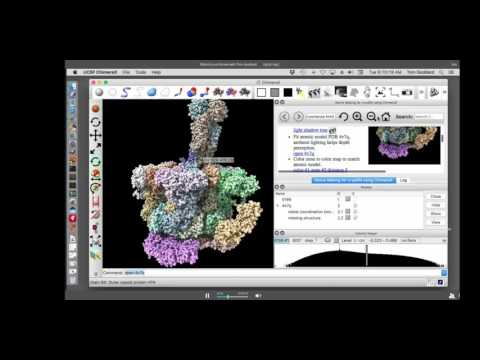
Topic: Movie Making for cryoEM using ChimeraX
Presenter: Tom Goddard, Programmer/Analyst, UCSF Resource for Biocomputing, Visualization, and Informatics
Hosted by Pete Meyer
Recorded on April 11, 2017

Topic: What's New in Coot?
Presenter: Paul Emsley, Ph.D., MRC Laboratory of Molecular Biology
Recorded on February 16, 2021
Host: Jason Key
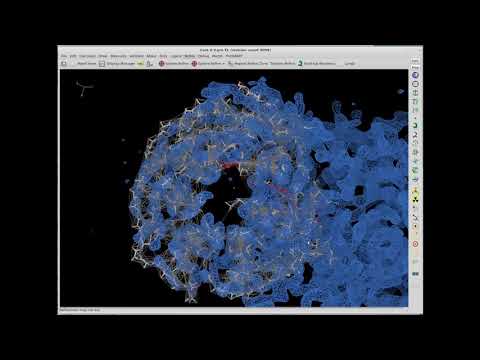
Topic: Breezing Through the Coot Cryo-EM Tutorial
Speaker: Paul Emsley - MRC Laboratory of Molecular Biology
Recorded March 2019
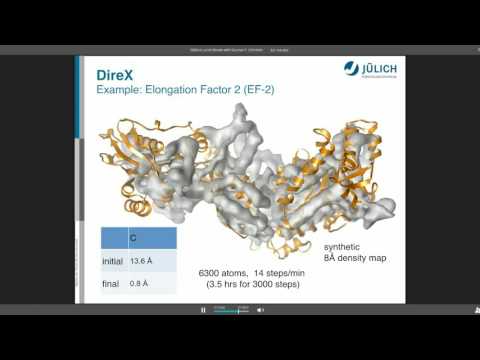
Topic: DireX - Cross-validated Real-space Refinement to Density Map
Presenter: Gunnar Schröder, PhD, Assistant Professor, University of Düsseldorf, Jülich, Germany
Host: Pete Meyer
Recorded on May 10, 2016
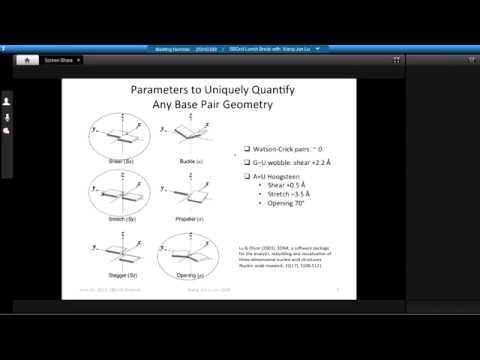
Topic: DSSR, a new 3DNA program for Defining the Secondary Structures of RNA from three-dimensional coordinates
Presenter: Xiang-Jun Lu, Ph.D., Associate Research Scientist, Columbia University
Host: Jason Key
Recorded on June 26, 2014
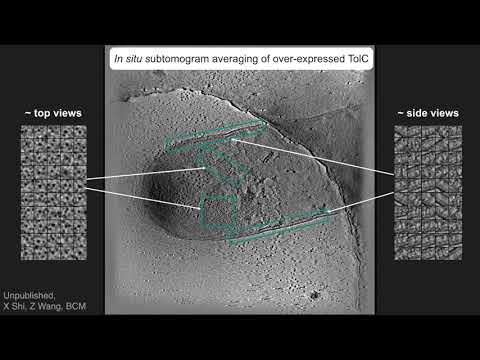
Topic: In-situ and in-vitro subtomogram averaging to subnanometer resolution using EMAN2.3
Presenter: Steven J Ludtke, Ph.D., Professor, Baylor College of Medicine
Recorded on November 12, 2019
Host: Jason Key

Topic: EMAN2
Presenter: Steven Ludtke, Ph.D. Baylor College of Medicine
Host: Jason Key
Recorded on May 11, 2021
Topic: A quick introduction to EMAN2.1 for CryoEM Single Particle Analysis
Presenter: Steven Ludtke, Ph.D., Professor, Baylor College of Medicine
Host: Jason Key
Recorded on April 15, 2015
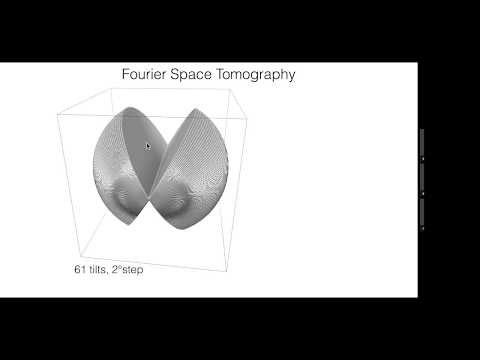
Topic: EMAN2.2 for Single Particle Analysis and In-situ Structural Biology
Presenter: Steven Ludtke, Ph.D.
Professor of Biochemistry - Baylor College of Medicine
Co-Director - National Center for Macromolecular Imaging
Co-Director - Center for Computational and Integrative Biomedical Research
Date Recorded: November 14, 2017
Host: Jason Key
Topic: EVcouplings: Using evolution to determine structure and function
Presenters: Kelly Brock, Ph.D., Laboratory of Debora Marks, Harvard Medical School and Thomas Hopf, Ph.D., Scientific Consultant
Host: Pete Meyer
Recorded on May 12, 2020
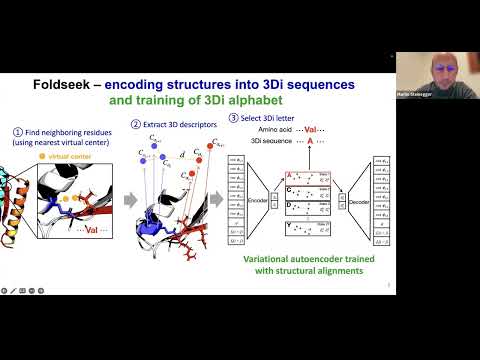
Topic: Fast and accurate protein structure search with Foldseek
Presenter: Prof. Martin Steinegger, Asst Professor of Bioinformatics, Seoul National University
Host: Pete Meyer
Recorded on Feb 14, 2023
For more information on Foldseek: https://github.com/steineggerlab/foldseek

Topic: Haddock
Presenter: Prof. Alexandre Bonvin, University Utrecht
Host: Jason Key
Recorded on: June 29, 2021
Topic: Modelling biomolecular complexes using HADDOCK: local vs server mode.
Presenter: Alexandre Bonvin, Computational Structural Biology group, Bijvoet Center for Biomolecular Research, University of Utrecht, The Netherlands
Host: Jason Key
Recorded on September 8, 2015
Topic: Creating and analyzing multiple sequence alignments with the Jalview Desktop Application
Presenter: Jim Procter, Ph.D., Jalview Coordinator, College of Life Sciences, University of Dundee Scotland, UK
Host: Jason Key
Recorded on July 11, 2013
Topic: Retraining AlphaFold2 yields new insights into its learning mechanism and capacity for generalization.
Presenter: Gustaf Ahdritz, Ph.D. Student, AlQuraishi Laboratory Columbia University.
Host: Jason Key
Recorded on April 11, 2023
For more information on OpenFold:
https://sbgrid.org/software/titles/openfold
https://github.com/aqlaboratory/openfold

Topic: OpenMM
Speaker: Peter Eastman, Senior Software Engineer, Vijay Pande's Group, Stanford University
Host: Jason Key
Recorded on February 28, 2017




















