Webinars
- Page 1 of 1
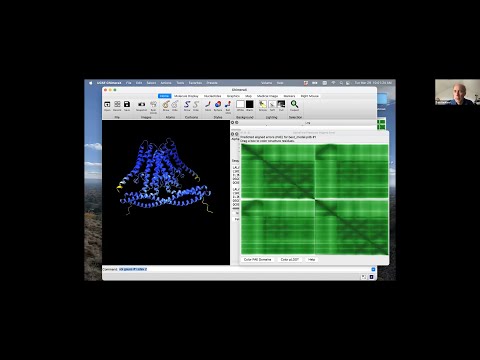
Presenter: Tom Goddard, UCSF Resource for Biocomputing, Visualization, and Informatics - Topic: Using AlphaFold protein structures in ChimeraX for cryoEM modeling
Linked to presented materials: https://www.rbvi.ucsf.edu/chimerax/data/sbgrid-mar2022/alphafold_pae.html
Learn more on the ChimeraX website:
https://www.rbvi.ucsf.edu/chimerax/data/alphafold-nov2021/af_sbgrid.html
This talk was presented as part of the SBGrid Spring Mini-series - Cryo-electron microscopy of membrane proteins: from sample to structure. See the full series lineup at https://sbgrid.org/webinars/
Organized by
Prof. Jamaine Davis, Meharry Medical College
Prof. Piotr Sliz, Harvard Medical School
Prof. Patrick Sexton, ARC Centre for Cryo-electron Microscopy of Membrane Proteins, Monash University
Recorded on March 29, 2022
SBGrid webinars are hosted with partial support from the NIH R25 Continuing Education for Structural Biology Mentors #GM151273, in collaboration with Co-PI Jamaine Davis of Meharry Medical College.
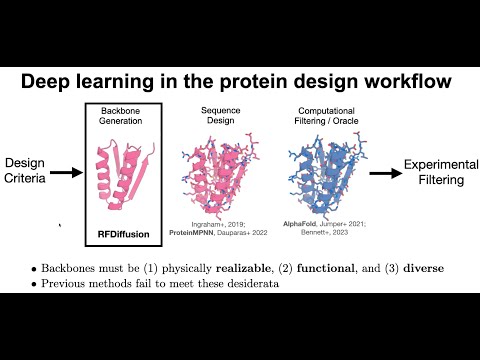
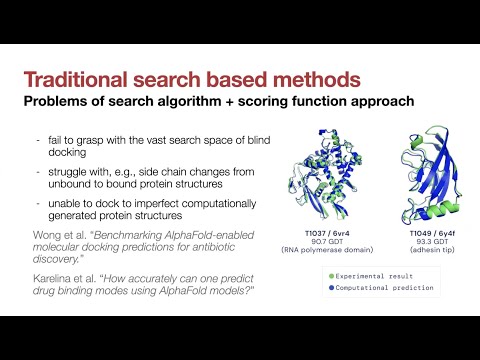
Topic: De novo design of protein structure and function with RFdiffusion.
Presenter: Brian Trippe, Ph.D., Postdoctoral Fellow, Columbia University.
Host: Pete Meyer
Recorded on April 9, 2024
For more information on RFdiffusion:
https://sbgrid.org/software/titles/rfdiffusion
https://github.com/RosettaCommons/RFdiffusion
SBGrid webinars are hosted with partial support from the NIH R25 Continuing Education for Structural Biology Mentors #GM151273, in collaboration with Co-PI Jamaine Davis of Meharry Medical College.
Topic: DiffDock: a state-of-the-art method for molecular docking.
Presenter: Gabriele Corso, Hannes Staerk, and Bowen Jing, Ph.D. students, MIT Computer Science and Artificial Intelligence Laboratory (CSAIL).
Host: Pete Meyer
Recorded on November 14, 2023
For more information on DiffDock:
https://sbgrid.org/software/titles/diffdock
https://github.com/gcorso/DiffDock

"Recent Enhancements and Scientific Advancements in the Schrödinger Suite"
Presenter: Woody Sherman, Schrödinger Vice President of Applications Science.
SBGrid Host: Piotr Sliz
Recorded on: June 12, 2012
Schrödinger website: http://www.schrodinger.com
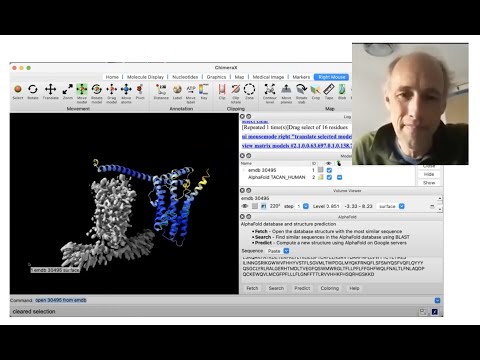
Topic: Using AlphaFold protein structures in ChimeraX for cryoEM modeling
Presenter: Tom Goddard, UCSF Resource for Biocomputing, Visualization, and Informatics
Q&A session also includes Shaun Rawson, who presented during this session on "Effective on-the-fly and downstream processing of CryoEM data." See https://youtu.be/ra2sVFEPtN8
Learn more on the ChimeraX website:
https://www.rbvi.ucsf.edu/chimerax/data/alphafold-nov2021/af_sbgrid.html
This talk was presented as part of the SBGrid Australasian III Mini-series - CryoEM: from Sample to Structure and should appeal to both novice and expert structural biologists.
See the full mini-series lineup at https://sbgrid.org/news/sbgrid-university-otago-webinar-series.