Lecture 17: Cctbx.xfel: New software for serial crystallography

Topic: Cctbx.xfel: New software for serial crystallography
Presenter: Nicholas Sauter, Lawrence Berkeley National Laboratory
Presented as part of:
SBGrid/NE-CAT 2014: Data Processing in Crystallography
June 5-6, 2014
Lecture 2: Scaling and Merging

Topic: Scaling and Merging
Presenter: Zbyszek Otwinowski, Professor, UT Southwestern Medical Center
Presented as part of:
SBGrid/NE-CAT 2014: Data Processing in Crystallography
June 5-6, 2014
Lecture 3: CC* - Linking crystallographic model and data quality

Topic: CC* - Linking crystallographic model and data quality
Presenter: Kay Diederichs, Professor, University of Konstanz
Presented as part of:
SBGrid/NE-CAT 2014: Data Processing in Crystallography
June 5-6, 2014
Lecture 6: MOSFLM and FRIENDS: Data Processing in the CCP4 suite.
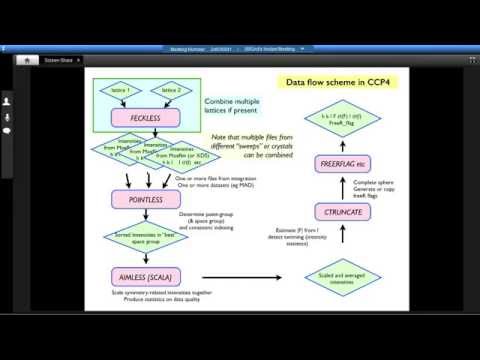
Presenter: Phil Evans, MRC Laboratory of Molecular Biology
Topic: MOSFLM and FRIENDS: Data Processing in the CCP4 Suite
SBGrid/NE-CAT 2014: Data Processing in Crystallography
June 5-6, 2014
Topic: HKL3000
Presenter: Zbyszek Otwinowski, UT Southwestern Medical School
Presented as part of:
SBGrid/NE-CAT 2014: Data Processing in Crystallography
June 5-6, 2014
Lecture 8: Principles of data processing with XDS

Presenter: Kay Diederichs, University of Konstanz, Germany
Topic: Principles of data processing with XDS.
Presented as part of:
SBGrid/NE-CAT 2014: Data Processing in Crystallography
June 5-6, 2014
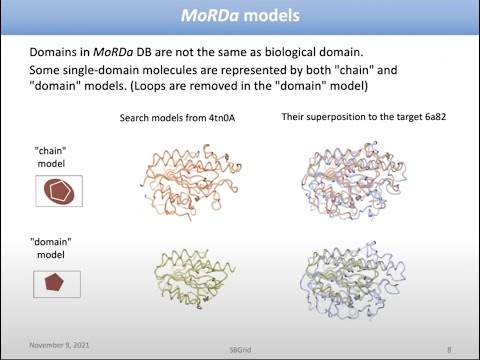
Topic: MoRDa, an automatic molecular replacement pipeline
Presenter: Andrey Lebedev, Ph.D., Science and Technology Facilities Council, UK
Host: Pete Meyer
Recorded on November 9, 2021
MovableType Software for Fast Free Energy-based Virtual Screening

Topic: MovableType Software for Fast Free Energy-based Virtual Screening
Presenter: Lance Westerhoff, Ph.D., QuantumBio, Inc.
Host: Jason Key
Recorded on October 6, 2020

Topic: PanDDA: extracting ligand-bound protein states from conventionally uninterpretable crystallographic electron density
Presenter: Nicholas Pearce, Postdoctoral Researcher, Utrecht University, The Netherlands
Recorded on: December 4th, 2018
Host: Jason Key
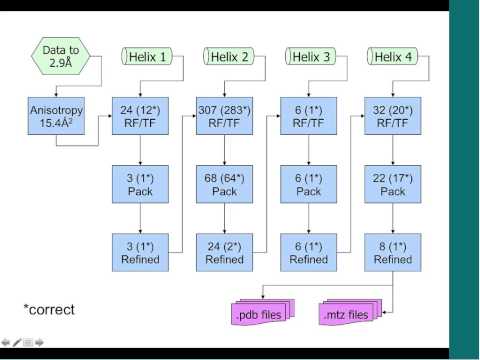
Topic: Extending the reach of molecular replacement in Phaser
Presenter: Randy Read, Wellcome Trust Principal Research Fellow in the Department of Haematology at University of Cambridge.
Host: Piotr Sliz
Recorded on April 23, 2012

Topic: Density Modification and Model Building with cryo-EM maps.
Presenter: Tom Terwilliger, Laboratory Fellow, Los Alamos National Laboratory
Recorded on March 31, 2020
Host: Jason Key
Link to slides: https://drive.google.com/open?id=1SHQ...
Topic: Atomic model refinement and validation in Phenix
Presenter: Pavel Afonine, Computational Research Scientist, Berkeley Lab
Date recorded: April 3, 2020
Host: Pete Meyer
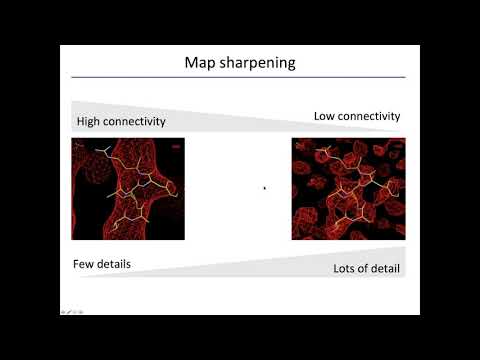
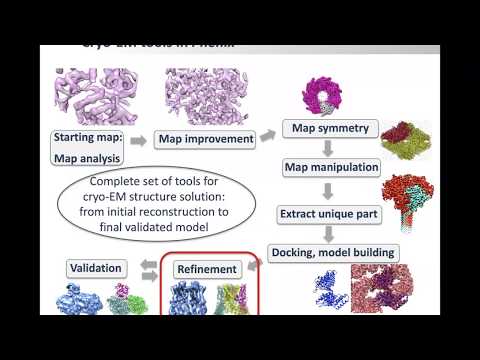
Topic: Map Sharpening; Model Building; other cryo-EM tools in Phenix
Presenter: Dorothee Liebschner, Berkeley Lab
Hosted by Jason Key
Recorded on April 7, 2020

Topic: Atomic model, map and model-to-map fit validation
Part I: Atomic model validation
Speaker: Pavel Afonine, Berkeley Lab
Host: Jason Key
Recorded on April 14, 2020
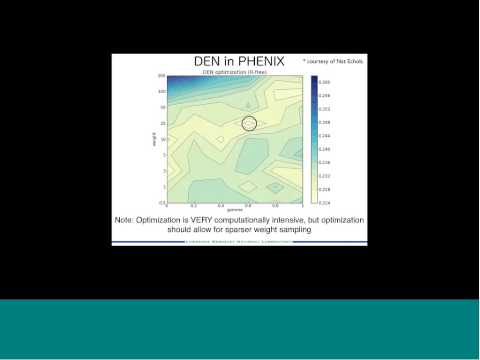
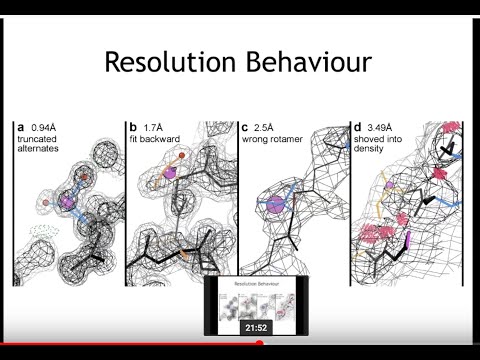
Topic: Phenix: Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution
Presenter: Jeff Headd, Postdoctoral Fellow at Lawrence Berkeley National Laboratories
Host: Piotr Sliz
Recorded on December 12, 2011
Topic: Improved chemistry restraints for crystallographic refinement by integrating the Amber force field into Phenix
Presenter: Nigel Moriarty, Lawrence Berkeley National Labs
Host: Pete Meyer
Recorded on: October 8, 2019

SBGrid webinars are hosted with partial support from the NIH R25 Continuing Education for Structural Biology Mentors #GM151273, in collaboration with Co-PI Jamaine Davis of Meharry Medical College.
Topic: AQuaRef - Machine Learning Accelerated Quantum Refinement of Protein Structures
Presenter: Pavel Afonine, Ph.D. Computational Research Scientist, Berkeley Lab
Host: Peter Meyer
Recorded on November 12, 2024
For more information on AQuaRef:
https://phenix-online.org/version_docs/dev-5412/reference/AQuaRef.html
https://github.com/qrefine/qrefine
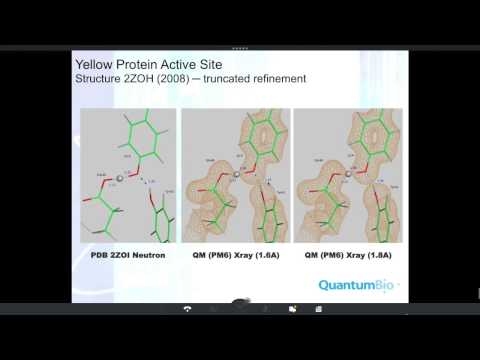
Topic: Phenix/DivCon: accurate macromolecular crystallographic refinement using linear scaling, semiempirical quantum-mechanics.
Presenter: Lance Westerhoff, President and General Manager
Host: Jason Key
Recorded on September 22, 2015
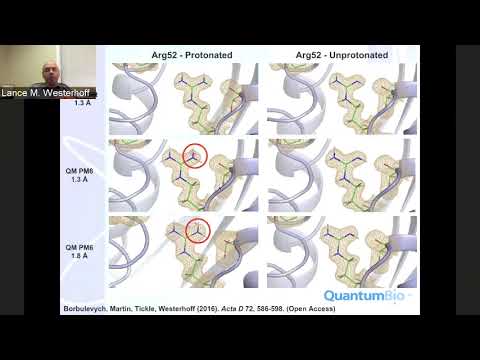
Topic: XModeScore: using crystallography to determine protonation states, fragment binding modes, and flip-states.
Speaker: Lance Westerhoff, President and General Manager at QuantumBio Inc.
Date: recorded: Oct 2, 2018
Host: Jason Key
Phenix-Rosetta Refinement
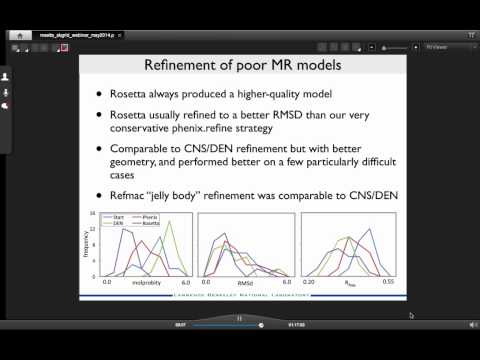
Topic: Refinement of challenging structures with Rosetta and Phenix
Presenter: Nat Echols, Computational Scientist at Lawrence Berkeley National Labs
Host: Andrew Morin
Recorded on May 13th, 2014




















