NE-CAT is pleased to offer our Practical Crystallography Course, June 21-23, 2023 at Harvard Medical School in Boston, Massachusetts. The course will be a three day workshop hosted in conjunction with SBGrid. We are currently planning for three full days with a break for lunch and potentially an afternoon before the start of the workshop to make sure that computers are properly setup. Lunch and coffee breaks will be provided. The workshop remains free, but students are responsible for their own lodging and transportation. All students will need access to SBGrid during the workshop in order to participate. Temporary access to SBGrid can be provided. The workshop is limited to 20 participants. Priority will be given to those with data and/or problematic datasets, but curated datasets are also …
SBGrid Consortium - News and Events
NE-CAT Practical Crystallography Course
Published March 23 2023
Animation Workshop with Janet Iwasa
Published June 7 2022
August 1-5, 2022
Boston MA
Organizers:
Prof. Janet Iwasa, The Animation Lab, University of Utah
Prof. Piotr Sliz, SBGrid, Harvard Medical School
SBGrid and Janet Iwasa of The Animation Lab are pleased to host a 5-day intensive, in-person workshop from August 1-5, 2022 that will cover 3D molecular animation. Students will learn how to export molecular models from UCSF Chimera, work together on a common project to create an animated scene in the 3D animation software Autodesk Maya, and use Adobe After Effects to composite the final animation, adding narration and text. Dr. Iwasa will reserve part of each day for students to ask questions about their specific planned animation with a goal that they will return to the laboratory …
SBGrid presentation at USNC/Cr
Published March 7 2022
Tuesday, April 5th, 11am-2pm EDT - Register here for the session
Hosted by the US National Committee for Crystallography, National Academies of Sciences, Engineering, and Medicine
In this session, attendees will learn
- How to use SBGrid Software Installer, AppCiter, and SBGrid Data Bank
- How to deploy SBGrid applications and experimental data on personal computers, HPC clusters, and in AWS.
- How to effectively utilize SBGrid applications in support of CryoEM, small-molecule docking and structure prediction.
- About the scientific software curation process - skills and expertise required to compile scientific software.
SBGrid Spring Webinar Mini-Series - CryoEM of membrane proteins: from sample to structure
Published February 24 2022
We're excited to announce a new webinar mini-series starting March 15th - CryoEM of membrane proteins: from sample to structure - with a terrific group of speakers including opening and closing keynotes from Profs. Nieng Yan and Chris Tate, a special lecture from Prof. Melanie Ohi, panel discussions on sample preparation and regional CryoEM infrastructure, and a look at Single Particle CryoEM with Shaun Rawson and ChimeraX CryoEM modeling with Tom Goddard.
See the full lineup and registration details at sbgrid.org/webinars/
Organized in collaboration with Profs. Jamaine Davis of Meharry Medical College, Piotr Sliz of Harvard Medical School, and Patrick Sexton at the ARC Centre for Cryo-electron Microscopy of Membrane Proteins, Monash University.
BioGrids for SBGrid Users Tutorial
Published November 5 2021
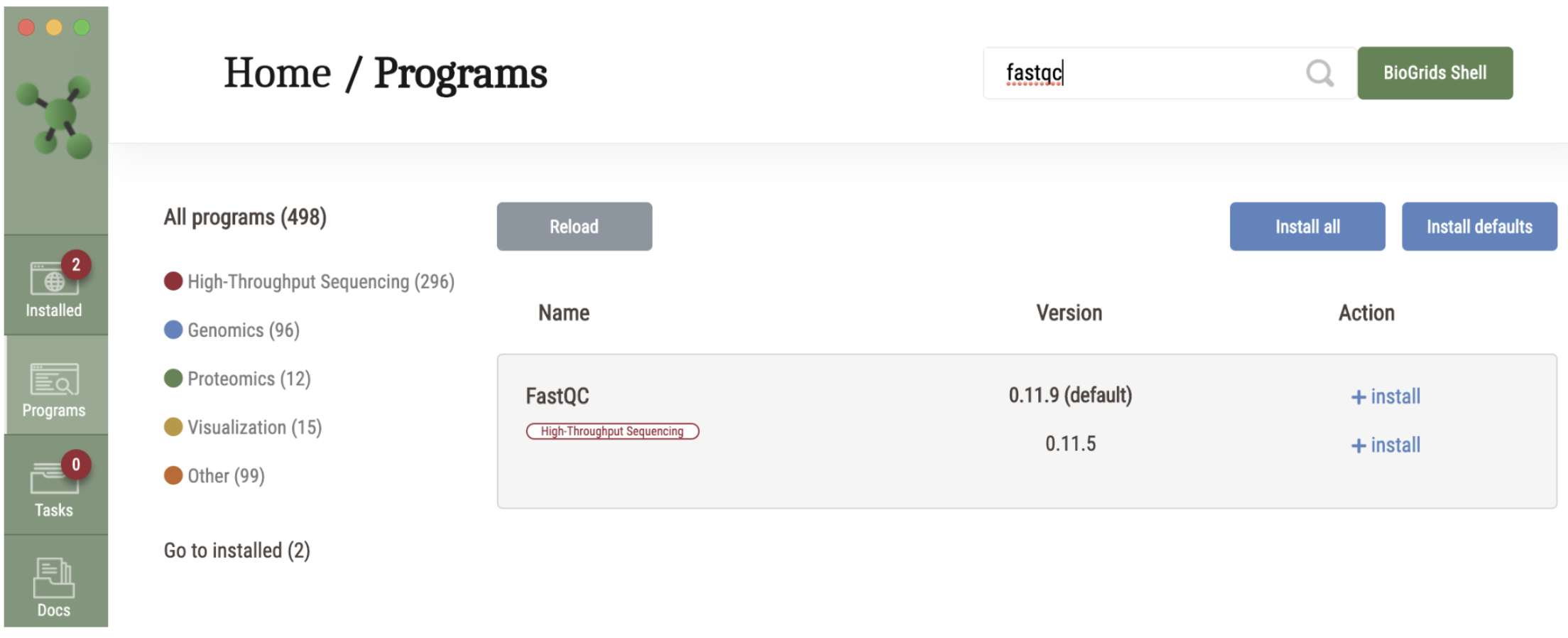
For SBGrid users who are already familiar with the SBGrid software installation platform, members may opt-in in to include the BioGrids software stack in their installation. BioGrids in an SBGrid-style collection of ~ 500 open source bioinformatics software titles, with collections for high-throughput sequencing, genomics, proteomics, visualization, image analysis, etc.
Our resident bioinformatics expert, Jim Vincent, recorded this brief tutorial - BioGrids for SBGrid Users - with more information on how SBGrid users can access this software stack. Contact us at help@biogrids.org with any questions.