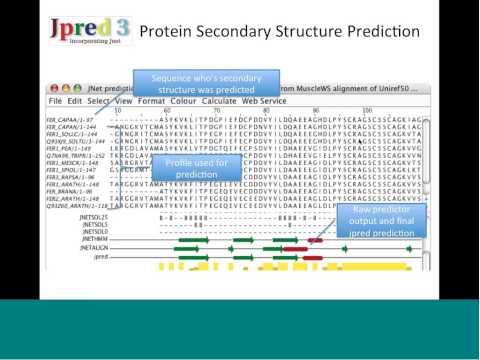
Into Alignment
In 1987, when Geoff Barton was a graduate student learning computational structural biology at the University of London, just 6000 protein sequences were known, but their numbers were rising exponentially, and it was becoming clear that they had commonalities. Sequences that yield valuable functions have staying power, so they are conserved throughout evolution. Finding these recurring patterns, however, required painstaking pencil and paper comparisons.