Computing Cellular Clockworks
Physicist Klaus Schulten once imagined becoming a dancer, relying on nothing but his own mind and body to perform. “But I was not a good dancer,” he says. “So my next thing was theoretical physicist. Just myself, pencil and paper — and in my case, also an eraser.”
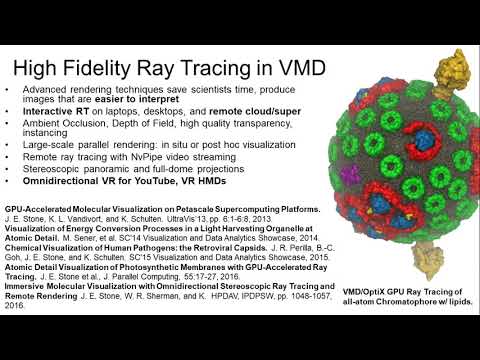
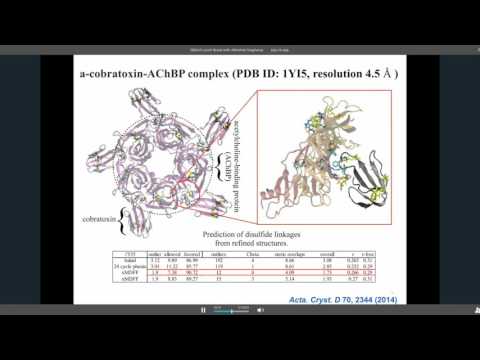
That dream was also thwarted. Today, Schulten relies on some of the most powerful and expensive computing equipment on earth to carry out his work, which applies theoretical physics to the understanding of biological systems. His most recent work involved the molecular simulation of an organelle that converts light energy to chemical energy inside a photosynthetic bacterium using the molecular dynamics tools NAMD and VMD, which his lab develops. Looking forward, he wants to simulate an entire living cell, right …
Read the full story here.