Tooling Around
Andrew Kruse
Harvard Medical School
Published March 29, 2018
Until recently, a vaccinated llama has been a membrane protein crystallographer’s best friend. That was before Andrew Kruse and his co-authors showed that yeast can be a faster, cheaper, and possibly better tool for otherwise impossible crystallographic studies.
Broadly, the Kruse lab at Harvard Medical School is interested in how cells transfer information across their membranes. To probe the finer points of the molecular interactions, Kruse and his collaborators have developed new tools, including the yeast platform and a new evolutionary approach, to make studies of these membrane proteins easier for themselves and others.
About 20 years ago, it was discovered that the unique immune systems of camelids (llamas, camels, and alpacas) produce a pared down version of antibodies, featuring a single binding domain called a nanobody. Nanobodies were a boon to structural biologists. Among their many uses, these small molecules stabilize membrane proteins in specific different active conformations for crystallization and detailed structural analysis.
Yet despite the transformative impact on structural studies, it takes months to produce specifically tailored nanobodies by llama immunization. It can be prohibitively expensive, and the particles might not work. As a junior faculty member, Kruse felt constrained by cost and time limitations of immunization. So did a fellow junior colleague, Aashish Manglik at University of California, San Francisco. Together, the labs came up with a solution.
The yeast project arose out of their need for a better way to stabilize G protein-coupled receptors. GPCRs are a proteins lodged in fatty cell membranes that are crucial to human health and disease. The receptors detect hormones and neurotransmitters and then signal other proteins in the cell to respond. Millions of people take drugs targeting GPCRs, such as antihistamines, bronchodilators, antihypertensive agents, and many others. Better understanding of the structure and interactions could help researchers design more effective drugs with fewer side effects.
The yeast platform is one step toward the next stage Kruse envisions for GPCRs: A rational approach to finding and designing drugs to trigger or block specific activity.
“In signal transduction, there’s never been a more exciting time,” he says. GPCRs have both greasy and water-soluble parts, making them difficult to study at the atomic level until recently. The first high-resolution structure of a human GPCR wasn’t solved until the year 2007, Kruse says, but now dozens have been probed in their many different active shapes.
To boost those efforts, Kruse and his collaborators have made the vials of specially engineered yeast and instructions freely available to others for non-profit research. They fielded more than 100 requests from researchers in 17 countries little more than a month after publishing February 12, 2018, online in Nature Structural & Molecular Biology. About half of those requests came from posting the paper on the preprint service, BioRxiv. Other labs are using the nanobodies for projects ranging from identifying inhibitors of the zika virus to detecting bacterial species to classical genetic screens.

For as long as Kruse can remember, he dreamed of becoming a scientist. He grew up in the Twin Cities, Minnesota. Kruse discovered structural biology as an undergraduate at University of Minnesota, as a double major in biochemistry and math. It was a satisfying blend of fundamental quantitative biology with direct applications to human health. He worked in the lab of Douglas Ohlendorf on the structures of bacterial toxins.
Working in the lab also helped open up another part of the world. Because of its growing importance in science, Kruse decided to learn a Chinese language. A lab administrator connected Kruse to a university in northeast China where she had worked for years before settling in Minnesota. Before going to graduate school at Stanford, Kruse spent a summer working at the school in China. There, he met his wife. They speak mandarin exclusively at home. He has no formal collaborations in China, but an acquaintance at the National Center for Protein Science in Shanghai is helping distribute his new nanobody yeast platform within the country.Kruse began working on GPCRs in graduate school in the Stanford University laboratory of Brian Kobilka. About halfway through Kruse’s PhD training, Kobilka shared the 2012 Nobel Prize in Chemistry with Robert Lefkowitz of Duke University for their discoveries on GPCRs.
In Kobilka’s lab, Kruse worked on a GPCR group called muscarinic acetylcholine receptors. In studies to understand the molecular basis for therapeutic drug action, he showed how tiotropium bromide (Spiriva), an inhaled drug for chronic obstructive pulmonary disease and for uncontrolled asthma, recognizes and blocks the muscarinic 3 (M3) receptor. The once-daily dose is effective in dilating bronchial airways for 24 hours, because the receptor closes around and locks onto the drug, trapping it in place, he found.
He also worked on the related M2 muscarinic receptor, showing how multiple compounds could bind simultaneously to the same receptor. In other projects, he and his Stanford co-authors developed a way to generate more selective affinity reagents for transmembrane proteins. The new method led to a patent and then to a company. In 2016, Kruse was selected by Forbes magazine for its 30-under-30 in health care, along with Aaron Ring, now at Yale University, who together had founded Ab Initio Biotherapeutics, Inc. Ring also collaborated with Kruse and Manglik, another co-founder, on the new yeast platform.
At Harvard, the Kruse lab continues to work on membrane signaling with a variety of approaches, both old and new. For example, they collaborated with another Harvard lab on a mass spectrometry technique to track signaling in living cells.
For a 2017 PNAS paper, they reached back a century to the days when medical researchers used material from nearby slaughterhouses. With a modern twist, they used mail-order frozen calf livers to track down the identity of Sigma 2, a receptor implicated in Alzheimer’s disease, schizophrenia and cancer. Despite its potential therapeutic importance, it had not been cloned. In this case, the lab purified the receptor from homogenized calf livers and revealed its identity as the transmembrane protein (TMEM) 97. They are working on solving the structure. Sigma receptors were discovered in the 1970s because of their response to analogs of drugs that activate opioid receptors. In 2016, the Kruse lab determined the structure of Sigma 1, an unusual receptor that cross reacts with many drugs and has been linked to neurodegenerative diseases, addiction and pain.
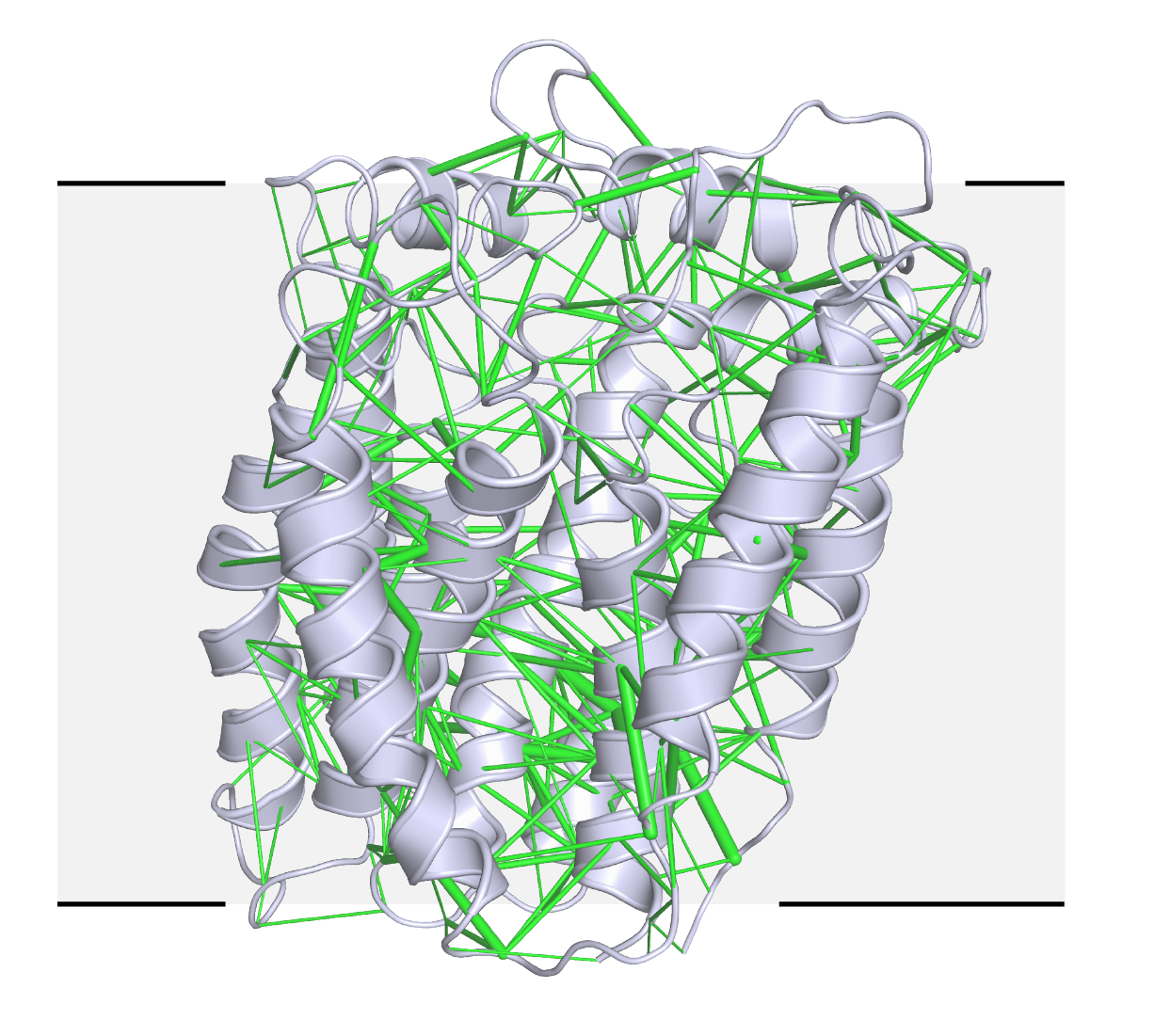
In a newer technique, they are using evolutionary analysis and prediction to solve structures from scratch, a collaboration with Harvard systems biologist Debora Marks. Structural biologists need a starting point to determine a new atomic structure, but there was no homologous protein for RodA, a transmembrane protein that bacteria use to build their cell walls. Evolutionary sequence data has been used to predict protein folds, but never to solve a crystal structure of a protein of this size, Kruse says. In this case, they generated hundreds of possible templates and tested them all. A few models were good enough to solve the structure (see image), published online 28 March in Nature. For the computational structure approach, they relied on molecular replacement software implemented by SBGrid, generating a shell script to iteratively run hundreds of searches.
--Carol Cruzan Morton





































































































































