Rules of motion
Priyamvada Acharya
Duke Human Vaccine Institute
Published July 31, 2020
Priyamvada Acharya studies how proteins on the surface of HIV move to invade cells and evade the immune system. For many years her research has been focused on the HIV-1 envelope (Env), a protein on the HIV-1 surface that the virus uses to enter human cells. In December 2019, she and her colleagues reported promising results of a new strategy that could lead to a long-sought vaccine.
Three months later, when the coronavirus pandemic was declared, the SARS-CoV-2 virus suddenly became an urgent target.
Acharya thought carefully about what her group at the Duke Human Vaccine Institute (DHVI) in North Carolina could contribute. Her team has been diving deeply into understanding the step-by-step shape changes of the HIV-1 Env that maneuver the virus into the host cell, including evasive moves that hide key regions from the immune system. They use that knowledge to design proteins to expose conformations to elicit a more effective immune response against the virus.
“This is what we know well,” she says. “We’re trying to bring everything we’ve learned from HIV into SARS, and apply the same rules of motion, and say, ‘Here is what we can to do to stabilize it in a form that potentially can lead to a vaccine.’”
Staying close to their roots has allowed Acharya’s team to move fast. “When you move to a new system, it’s hard,” she says. “If we started to learn from scratch, we were never going to contribute anything helpful in the time frame needed. It turned out that the underlying rules of protein structure and motion are the same for all proteins.”
Acharya, only two years into her new job as the Director of DVHI's Division of Structural Biology, tapped into her academic start-up funds for a fast pivot, while she and collaborator Rory Henderson, who heads Molecular Dynamics and simulation in DHVI Structural Biology, applied for more sustained funding for their new coronavirus research. Her lab was allowed to stay open to continue their COVID-19 research while most university research was locked down.
The scientific search for an HIV vaccine has taken more than three decades, but “with SARS-CoV-2, the barriers appear to be lower,” Acharya says. “You don’t really know until a vaccine is tested, but I’m optimistic. The simple example is that the receptor binding domain in SARS is well exposed, which is why there are all these antibodies that bind this functionally important region known as the receptor binding domain (RBD) and neutralize the virus. In HIV, on the other hand, the receptor binding regions are well hidden from the immune system.”
By the end of July 2020, Acharya’s team published five preprints in quick succession on the BioRxiv preprint server with a priority goal of sharing information with other scientists. The first paper, soon published in July Nature Structural and Molecular Biology demonstrated that the “up” and “down” RBD conformations of the SAR-CoV-2 spike can be controlled via structure-guided design. By tweaking the genetic sequence, her team generated soluble spike receptor binding domains locked into either the “up” or “down” position, providing a framework for rational design of potential vaccine candidates.
Next, their focus turned to the reactive antibodies triggered by the RBD. Found at the tips of the spike protein, the domain generates most of the antibodies measured in people infected with the coronavirus. The researchers wanted to explore other antibodies against the virus, so they designed a molecule with the protein locked in the “down” position.
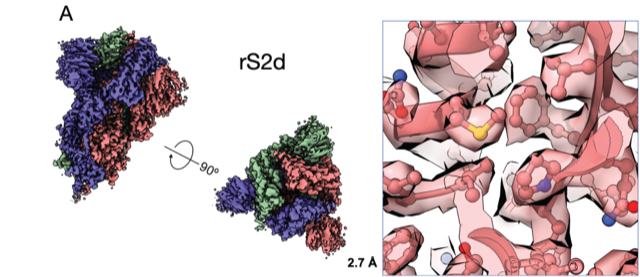
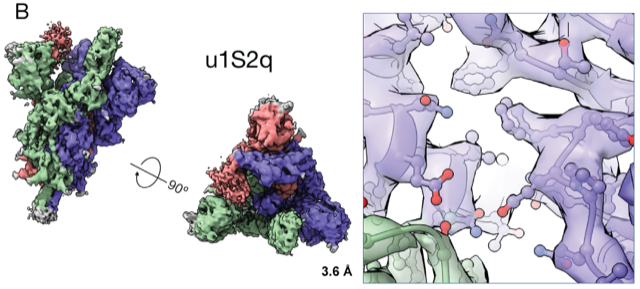
“We bring some of our fears and insecurities from the HIV field,” she says. “If something is immune dominant and elicits so many antibodies, the virus will eventually find a way to escape. One good thing is that SARS-CoV-2 is changing much slower than HIV, and the receptor binding domain is exposed and vulnerable.” In fact, they found, an early mutation in the virus seems to have made it more vulnerable to potential vaccines, they reported in another preprint.
Acharya’s team shares structural analysis with her DHVI colleagues in immunology and virology. They assess the new molecules in preclinical testing to see which ones work best to stimulate the immune system to block the various convolutions of viral infection. They also can use the designer molecules as probes to analyze antibodies in patient samples. Her team verifies promising molecular configurations. If all goes well, other colleagues can begin human trials. “It’s a continuous iterative process,” Acharya says.
As far back as she remembers, Acharya wanted to be a scientist. Acharya was born and raised in India, the daughter of an army architect (dad) and artist (mom). She calls no one place there home, because her family moved every two years, giving her a diverse and varied experience of her native country.
After concentrating on organic chemistry as an undergraduate and earning her PhD in biochemistry, she decided to pursue structural biology, because she wanted to see molecules move. After college, she relocated to the Max Planck Institute of Biophysics in Frankfurt, Germany, where she held a joint visiting scientist appointment with the Max Planck Institute for Terrestrial Microbiology, Marburg. Here, Acharya learned X-ray crystallography and worked on enzymes involved in methanogenesis and other biosynthetic pathways in archaebacteria.
In 2005, she took her new X-ray crystallography skills to the US National Institutes of Health to examine how HIV interacted with the host cell surface. In Peter Kwong’s lab, she studied how the HIV-1 Env proteins move when they first engaged with the CD4 receptor and then with CCR5, the final move before the virus fuses into the human cell.
In 2015, in a culmination of a lot of basic science work, she and her colleagues in the Kwong lab published the creation of a molecule in the HIV envelope fixed into a shape that prevented further movement and also attracted broadly neutralizing antibodies. The same year, Acharya moved to New York Structural Biology Center as an embedded scientist to incorporate electron microscopy studies into the work on HIV-1 entry and vaccine design. The cryo-EM studies in New York sped up the follow-up work.
In 2017, she and her colleagues showed what the mutated molecule was doing: It allowed CD4 to bind in the first step of HIV attachment, but blocked further conformational changes. Both papers were published in Nature Structural and Molecular Biology. In 2018, she and her colleagues used a similar approach to target HIV’s fusion protein. A molecule that locked its immunogenic area in an exposed state allowed elicitation of antibodies that targeted the HIV-1 fusion peptide. Acharya solved cryo-EM structures to visualize the epitopes of these fusion peptide-directed antibodies on HIV-1 Env. The paper, published in Nature Medicine, was promising evidence that a vaccine was possible.
The same year, Acharya was recruited to DHVI to continue to build a complete HIV vaccine. She embraced the highly collaborative bench-to-bedside environment with colleagues who can use the structural information to inform studies of viruses, their proteins, and immune responses in animals and people.
“Translation is very fast,” she says. “When you’re working that closely with a person, sometimes all you have to do is hand them a tube or give them an idea.”
Cryo-em has been key to understand the molecular mechanisms of both HIV and SARS and manipulate them for the good of human health, she says. Another powerful tool, called negative stain EM, run by Robert J Edwards, head of Negative Stain EM in DHVI Structural Biology, provides a quick assessment of structures at low resolution so the team can select candidates to evaluate further on cryo-EM grids.
Acharya has built most of her DHVI group from scratch. Early on, she looked for people who enjoyed and excelled at previous jobs, no matter the skill set and then gave them the space and various tasks to explore lab duties to see what strengths reveal themselves. For example, a nurse hired for administrative tasks is now one of the lab’s top biochemists.
In the lingering months of the pandemic, Acharya’s structural biology work is focused on the SARS-CoV-2 spike and the HIV-1 Env. For SARS, the major goals are to continue to understand the mechanics of the spike and translate this knowledge toward developing a pan-coronavirus vaccine in the long term. For the short term, Acharya’s team is working closely to support the DHVI vaccine teams by rapidly determining structures of new antibodies that will both be useful for passive vaccination and will also inform ongoing active vaccine studies.
For HIV-1, the group is using structural biology to trace the development and maturation of antibodies in HIV infected patients, and translating this knowledge to guide the immune system to develop a similar response. The first steps were taken in the December 2019 Science paper, where Acharya and her colleagues reported an engineered HIV-1 protein that stimulated and multiplied rare antibody responses in people. The next steps are to guide these responses to become broadly neutralizing antibodies against the virus.
-Carol Cruzan Morton





































































































































