Right place, right time
Ernesto Fuentes
University of Iowa College of Medicine
Published January 29, 2021
As a kid, Ernesto Fuentes lived in Brazil just long enough to stock up on the soccer skills he now shares with his kids. His affinity for structural biology may have arisen at the same time, shaped by family trips to see churches and other architectural highlights in nearby countries. Little did he know at the time, he would grow up to bring fresh insights to common building blocks responsible for a dazzling diversity of normal molecular function in living cells.
Back in the United States, Fuentes attended college in Ohio. He intended to become a chemical engineer, like his dad. Then he learned that proteins, like buildings, have distinctive architecture. He pivoted to a biology masters degree and followed up with a PhD specializing in nuclear magnetic resonance (NMR).
His doctoral studies immersed him in NMR and dynamics in more ways than one. His mentor’s lab moved twice, from University of Illinois to State University of New York Buffalo and then to University of Pennsylvania.
As a postdoctoral fellow in a University of North Carolina cancer laboratory, Fuentes established a paradigm for his future lab: Collaborate with colleagues doing the biology.
That leaves Fuentes free to explore fundamental questions about how a common part of many proteins can be involved in so many specialized tasks inside cells.
His postdoctoral work introduced Fuentes to PDZ domains, a small protein region found in more than 250 proteins that cover a broad swath of biology. PDZ domains connect partner proteins to each other in large signaling structures inside a cell next to the membrane in an arrangement that facilitates specific biological action.
PDZ domains somehow put together the protein scaffolding “in the right place at the right time” to get things done, Fuentes says.
Thirty years ago, scientists figured out the basic function: A PDZ domain in one protein grabs another protein by its tail. The tail, or C-terminus, fits neatly into the main PDZ groove. But there are subtleties in that maneuver that dictate exactly what other proteins can connect and what they can do as a result, Fuentes and others are discovering as they develop new ways to learn the rules of the PDZ family specificity.
Since Fuentes joined the University of Iowa College of Medicine faculty in 2006, his lab has been probing the specialized interactions of PDZ domains.
In their research, the Fuentes lab applies multiple integrated structural biology techniques for deeper insight, including NMR, X-ray scattering, crystallography, and cryo-electron microscopy, leaning on a full repertoire of SBGrid software.
“We don’t do biology, but we’re really interested in biology,” Fuentes says. “We’re developing tools that allow us to interrogate biology. We can do that through structure.”
The PDZ domain is found in many proteins and a protein can have one or many PDZ domains. Not only is PDZ a popular domain in proteins, it is a veritable social butterfly. PDZ domains bind to their counterparts in other proteins and mix it up with other ligands. Learning how finely tuned PDZ domains meet up with binding partners will illuminate normal biological signaling and may help develop more precise therapeutics.
PDZ domains help build protein complexes that facilitate different types of cell adhesion and signaling. In particular, they allow epithelial and neuronal cells to form cell-cell adhesion complexes that are critical for normal tissue and neuronal function, Fuentes says. Mutations that affect PDZ activity can have health consequences, including cancer, deafness, neuropsychiatric conditions, and cystic fibrosis.
In a central question, the lab has been asking how selective are PDZ domains in their binding partners, also known as specificity.
To sum up a dozen years of work with three PDZ family members (CASK, TIAM1/2, and Scribble), Fuentes and his colleagues have found that the PDZ groove for a protein tail is just a starting point. Some of the specificity comes from dynamics in the other PDZ nooks and crannies known as subpockets. Not only can certain amino acids wiggle into place better than others on a PDZ domain, the PDZ subpockets will adjust themselves to accommodate a preferred protein partner.
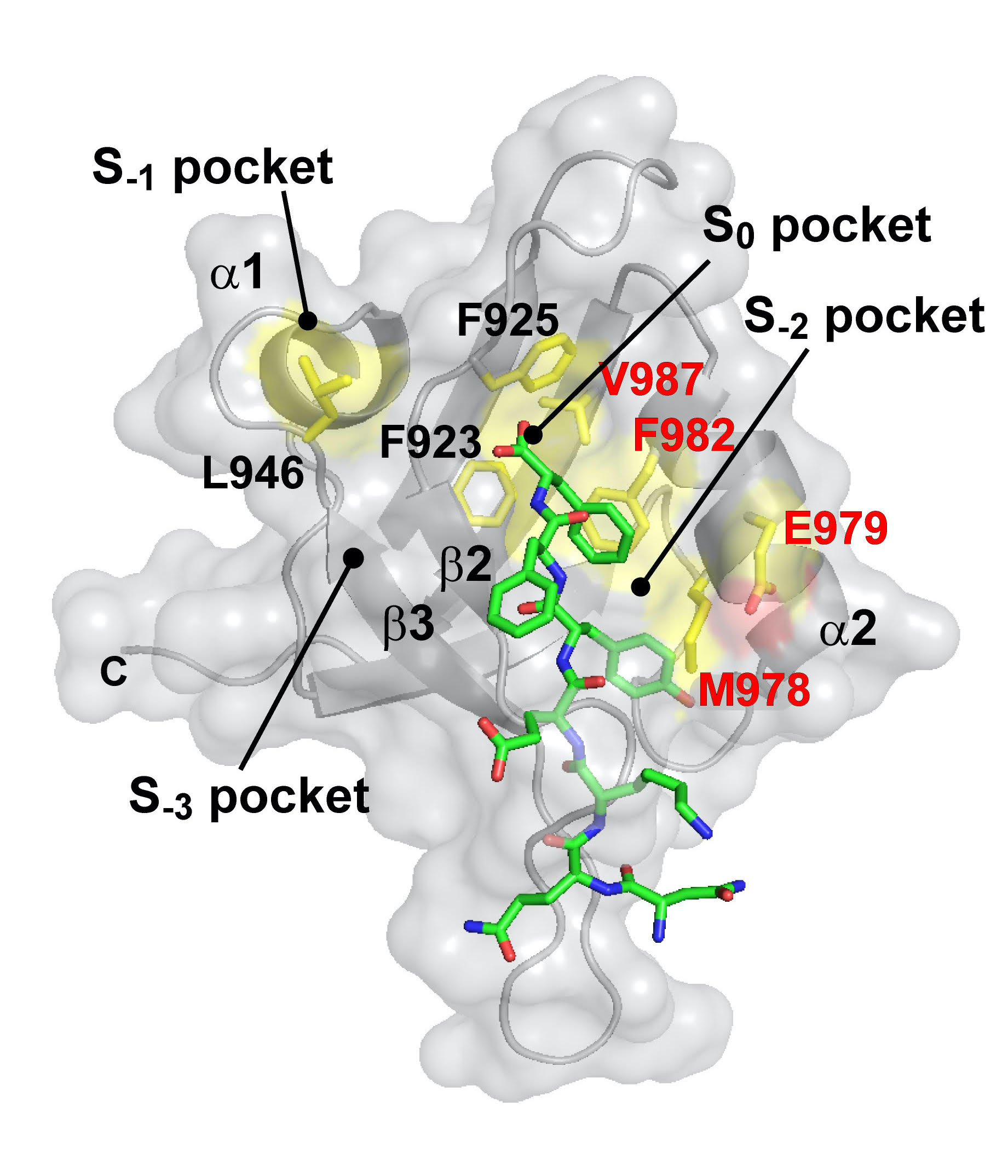
“Changes in the dynamics might be key to understanding specificity,” Fuentes says. For example, a protein with more dynamic range may be able to accommodate more partners and participate in different types of signaling. A less dynamic protein may have a more specific role. That means a mutation could allow more or less dynamic changes, influencing the biological system.
The researchers have experimented with mutations in different PDZ subpockets to observe the impact on binding. Some mutations sparked increased dynamics, also called hot spots, and those PDZ domains favored different or more binding partners.
“The really cool thing, this gave us insight into how evolution can change the specificity of a single PDZ domain,” Fuentes says. “Over time, mutations may be able to broaden or direct the domain to a new set of targets.”
In another twist, they found that some PDZ domains can bind internal motifs and not just the canonical C-terminal tail. “What’s interesting is that it opens up another set of interactions and set of proteins,” he says. “There are not that many examples, and the specificities are not well understood.”
Fuentes found a rare example in a collaboration with a cancer researcher who is studying how epithelial cells normally are held together so tightly. Most cancers originate in epithelial cells, and cancers have to overcome the cell-to-cell adhesion system to grow and spread.
In the project, Fuentes' team was tasked with dissecting an interaction between the proteins SGEF and Scribble involving a PDZ domain. They found an internal PDZ motif in SGEF that binds to one of Scribble’s PDZ domains.
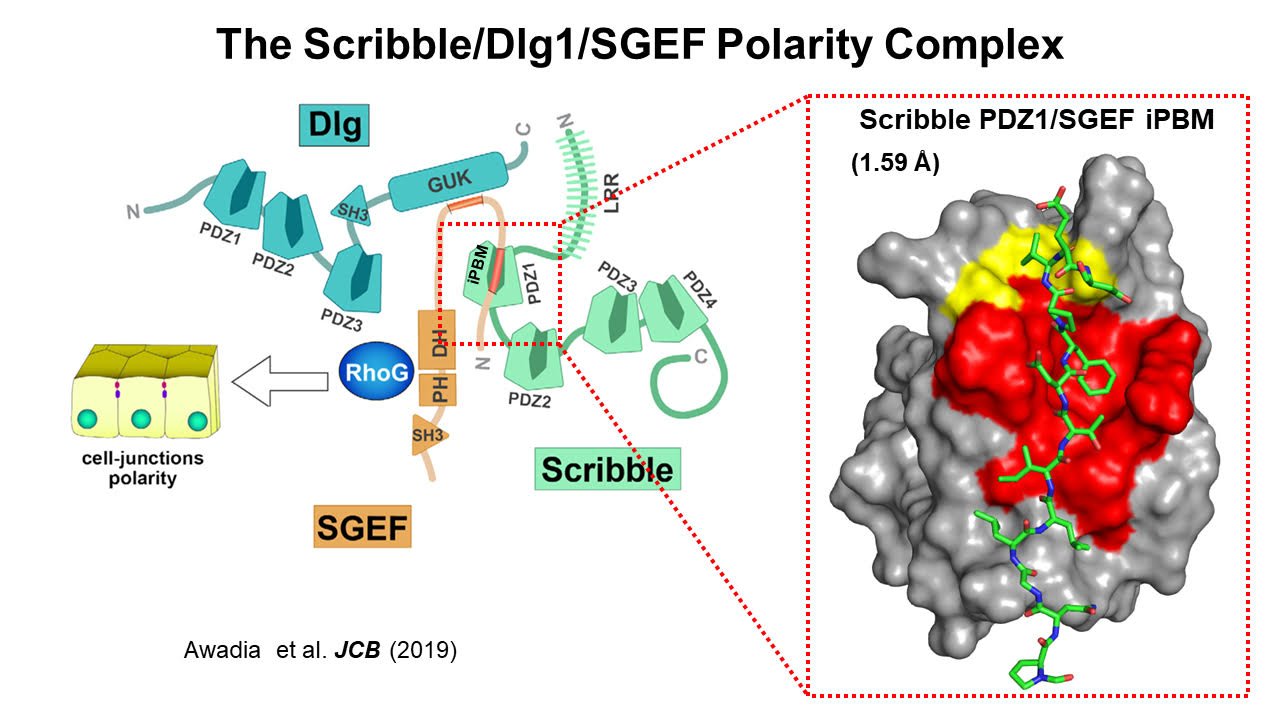
But a crystal structure revealed a new mechanism of PDZ specificity. The SGEF internal motif binds tightly because additional amino acids can latch on to an extra compatible section of Scribble’s PDF domain. The paper was published June 2019 in the Journal of Cell Biology with co-author Rafael Garcia-Mata at the University of Toledo.
The Fuentes lab will work toward a structure of the larger molecular complex with electron microscopy. But the experimental model also opens up a new way to study PDF specificity in more detail using synthetic biology methodologies in collaboration with a research team at Johns Hopkins University.
“We’re now looking at artificial designs of PDZ with the idea of obtaining a deeper understanding of the rules for how nature encodes specificity in a small domain,” Fuentes says. Preliminary work has produced a template protein domain with structural and biophysical properties of a PDZ. “We have a blank slate to design and test the features of specificity,” he says.
In other non-PDZ work from the Fuentes lab, they and their collaborators discovered details of the SrrB two-component signaling system that helps Staphylococcus aureus sense a change from an aerobic to an anaerobic environment.
S. aureus is a common bacterium found in people’s noses and on skin, but inside the body it can turn into a potentially deadly infection. Fuentes struck up a collaboration with a university colleague, Pat Schlievert, who discovered a two-component signaling system important for environmental sensing that allows S. aureus to make the dangerous adaptation to a low oxygen environment.
As they reported in a 2019 paper in PNAS, the Fuentes team found redox active cysteines in a sensor histidine kinase had a big influence on S. aureus pathogenicity.
If there is a theme running through the Fuentes lab, it’s to build an integrative structural biology approach using the tools that best suit the needs of the problem. And to keep an eye on the bigger picture – the biology.
“Structural biology is a part of many studies,” Fuentes says. “It’s not always the central feature. It’s an added method that allows deeper insight into the whole system.”
-Carol Cruzan Morton
Other tales
-
![]()
The Stories Antibodies Tell
Jean-Philippe Julien
Published 27 January 2026
![]()
Reshaping Membranes
Melanie Ohi
Published 23 November 2025
-
![]()
Probing Microbes
Gira Bhabha
Published 30 September 2025
![]()
Drawn to the Light
Emina Stojković
Published 30 July 2025
-
![]()
The Final Phase
George Phillips
Published 31 May 2025
![]()
Mind and Muscle
Ryan Hibbs
Published 28 March 2025
-
![]()
The Shapes of Energy
Luke Chao
Published 12 December 2024
![]()
Predicting Proteins
Jens Meiler
Published 25 November 2024
-
![]()
Death Metal
Steven Damo
Published 28 April 2024
![]()
Context Matters
Bing Chen
Published 30 January 2024
-
![]()
The Crystal Whisperer
Sarah Bowman
Published 29 November 2023
![]()
Data in Motion
Nozomi Ando
Published 29 September 2023
-
![]()
The Monstrous Maw
André Hoelz
Published 28 June 2023
![]()
Second Takes
Andrea Thorn
Published 28 February 2023
-
![]()
Radical reactions
Yvain Nicolet
Published 31 January 2023
![]()
Floppy Physics
Eva Nogales
Published 30 November 2022
-
![]()
Structure of Equity
Jamaine Davis
Published 28 September 2022
![]()
Life and Death of a Cell
Evris Gavathiotis
Published 28 July 2022
-
![]()
Follow the glow
Kurt Krause
Published 29 April 2022
![]()
Resolution solutions
Willy Wriggers
Published 25 February 2022
-
![]()
Of enzymes and membranes
Ming Zhou
Published 28 October 2021
![]()
Step-by-step
Gabrielle Rudenko
Published 26 September 2021
-
![]()
Moving muscle
Montserrat Samso
Published 26 July 2021
![]()
Particle catcher
Stefan Raunser
Published 28 June 2021
-
![]()
Designer drugs
Ho Leung Ng
Published 25 February 2021
![]()
Right place, right time
Ernesto Fuentes
Published 29 January 2021
-
![]()
Shape-shifting secrets of membranes
James Hurley
Published 27 November 2020
![]()
Enzymatic action
Cynthia Wolberger
Published 28 September 2020
-
![]()
Rules of motion
Priyamvada Acharya
Published 31 July 2020
![]()
Cosmic Squared
Michael Cianfrocco
Published 27 June 2020
-
![]()
Kaps are Cool
Yuh Min Chook
Published 28 April 2020
![]()
Spiraling into focus
Carsten Sachse
Published 29 March 2020
-
![]()
Seeing cilia
Alan Brown
Published 27 February 2020
![]()
For the Love of EM
Guy Schoehn
Published 27 January 2020
-
![]()
Protein Puddles
Michael Rosen
Published 16 December 2019
![]()
Changing channels
Daniel Minor Jr.
Published 27 September 2019
-
![]()
Listening Tips
Marcos Sotomayor
Published 30 July 2019
![]()
Beyond Cool
Published 31 May 2019
-
![]()
Hao Wu
A Higher Order
Published 30 May 2019
![]()
Aye Aye Captain
Alexandre Bonvin
Published 29 April 2019
-
![]()
The PARP Family Family
John Pascal
Published 28 February 2019
![]()
Frame by frame
Nikolaus Grigorieff
Published 28 January 2019
-
![]()
Predicting Success
Bil Clemons
Published 18 December 2018
![]()
Curiouser and Curiouser
Ramaswamy Subramanian
Published 27 November 2018
-
![]()
Rely on This
Sjors Scheres
Published 26 October 2018
![]()
Proteins out of bounds
Gerhard Wagner
Published 27 September 2018
-
![]()
Hiding in plain sight
Gaya Amarasinghe
Published 27 July 2018
![]()
Jumping Genes
Orsolya Barabas
Published 27 June 2018
-
![]()
Data Whisperer
Karolin Luger
Published 30 May 2018
![]()
Flipping the Switch
Jacqueline Cherfils
Published 27 April 2018
-
![]()
Tooling Around
Andrew Kruse
Published 29 March 2018
![]()
Comings and Goings
Tom Rapoport, Ph.D.
Published 23 February 2018
-
![]()
Transcriptional Rhythm
Seth Darst
Published 27 January 2018
![]()
The Language of Gene Regulation
Daniel Panne
Published 21 November 2017
-
![]()
Not Your Average Protein
James Fraser
Published 23 October 2017
![]()
Message Received
Sebastien Granier
Published 24 August 2017
-
![]()
Resistance is Futile
Celia Schiffer
Published 28 July 2017
![]()
Twist of Fate
Leemor Joshua-Tor
Published 28 June 2017
-
![]()
Drug Designer
John Buolamwini
Published 30 May 2017
![]()
Mathematically Minded
James Holton
Published 28 April 2017
-
![]()
Garbage Out
Kay Diederichs
Published 30 March 2017
![]()
Fixer Upper
Brandt Eichman
Published 27 February 2017
-
![]()
Mobilizers
Phoebe Rice
Published 31 January 2017
![]()
Escape Artist
Katya Heldwein
Published 19 December 2016
-
![]()
Nature’s Confectioner
Jochen Zimmer
Published 29 November 2016
![]()
State of Fusion
Jason McLellan
Published 27 October 2016
-
![]()
Here Be Dragons
Brian Fox
Published 28 September 2016
![]()
SBGrid Assumes Ownership of PyMOLWiki
Published 15 September 2016
-
![]()
Pharm Team
Oleg Tsodikov
Published 24 August 2016
![]()
Spiro-Gyra
Alejandro Buschiazzo
Published 27 July 2016
-
![]()
Turning the DIALS
Nicholas Sauter
Published 29 June 2016
![]()
Pipeline Dreams
Bridget Carragher and Clint Potter
Published 26 April 2016
-
![]()
U-Store-It
The SBGrid Data Bank provides an affordable and sustainable way to preserve and share structural biology data
Published 28 March 2016
![]()
Big Questions, Big Answers
Jennifer Doudna
Published 22 February 2016
-
![]()
Not a Structural Biologist
Enrico Di Cera
Published 17 December 2015
![]()
Divide and Conquer
Kevin Corbett
Published 19 November 2015
-
![]()
Computing Cellular Clockworks
Klaus Schulten
Published 23 October 2015
![]()
Trans-Plant
Gang Dong
Published 26 September 2015
-
![]()
Keep on Moving
James Berger
Published 23 August 2015
![]()
Totally Tubular
Antonina Roll-Mecak
Published 27 July 2015
-
![]()
From Disorder, Function
Julie Forman-Kay
Published 29 June 2015
![]()
Into Alignment
Geoff Barton
Published 27 May 2015
-
![]()
Two Labs, Many Methods
Michael Sattler
Published 28 April 2015
![]()
Picture This
Georgios Skiniotis
Published 20 March 2015
-
![]()
Intron Intrigue
Navtej Toor
Published 20 February 2015
![]()
Cut and Paste
Martin Jinek
Published 28 January 2015
-
![]()
Basics and Beyond
Qing Fan
Published 18 December 2014
![]()
Bloodletting and Other Studies
Pedro José Barbosa Pereira
Published 25 November 2014
-
![]()
Wire Models, Wired
A brief history of UCSF Chimera
Published 29 October 2014
![]()
In Search of…New Drugs
Doug Daniels
Published 30 September 2014
-
![]()
An Affinity for Affinity…and Corals
John C. Williams
Published 29 August 2014
![]()
Pete Meyer, Ph.D.
Research Computing Specialist
Published 22 August 2014
-
![]()
Justin O'Connor
Sr. System Administrator
Published 20 August 2014
![]()
Carol Herre
Software Release Engineer
Published 15 August 2014
-
![]()
Elizabeth Dougherty
Science Writer
Published 13 August 2014
![]()
Andrew Morin, Ph.D.
Policy Research Fellow
Published 11 August 2014
-
![]()
Jason Key, Ph.D.
Associate Director of Technology and Innovation
Published 8 August 2014
![]()
Piotr Sliz, Ph.D.
Principal Investigator, SBGrid
Published 1 August 2014
-
![]()
New Kid on the Block
James Chen
Published 29 July 2014
![]()
Membrane Master
Tamir Gonen
Published 30 June 2014
-
![]()
The Natural Bridge
Piotr Sliz
Published 13 June 2014
![]()
Surprise, Surprise
Catherine Drennan
Published 26 April 2014
-
![]()
Gone Viral
Olve Peersen
Published 20 March 2014
![]()
All Who Wander Are Not Lost
Frank Delaglio
Published 24 February 2014
-
![]()
The Raw and the Cooked
Graeme Winter
Published 24 January 2014
![]()
Vacc-elerator
Peter Kwong
Published 17 December 2013
-
![]()
Structural Storyteller
Karin Reinisch
Published 15 November 2013
![]()
The Fixer
Jane Richardson
Published 28 October 2013
-
![]()
Inside the Box
Mishtu Dey
Published 17 September 2013
![]()
Sensing a Change
Brian Crane
Published 16 August 2013
-
![]()
Towards Personalized Oncology
Mark Lemmon
Published 16 July 2013
![]()
Brush with Fame
Yizhi Jane Tao
Published 14 June 2013
-
![]()
Toxic Avenger
Borden Lacy
Published 21 May 2013
![]()
Pushing the Boundaries
Stephen Harrison
Published 22 April 2013
-
![]()
Strength in Numbers
Joseph Ho
Published 18 March 2013
![]()
One Lab, Many Methods
Wesley Sundquist
Published 12 February 2013
-
![]()
Unplanned Pioneer
Tim Stevens
Published 15 January 2013
![]()
From Actin to Action
Emil Pai
Published 11 January 2013
-
![]()
Unstructured
A Brief History of CCP4
Published 12 December 2012
![]()
Stop, Collaborate and Listen
Eleanor Dodson
Published 5 November 2012
-
![]()
X-PLORer
Axel Brunger
Published 1 October 2012
![]()
Share the Wealth
Zbyszek Otwinowski
Published 22 August 2012
-
![]()
Unraveling RNA
Anna Pyle
Published 18 July 2012
![]()
Sharper Image
Pawel Penczek and SPARX
Published 4 June 2012
-
![]()
Creative Copy Cat
Pamela Bjorkman
Published 25 April 2012
![]()
Charm and Diplomacy
Gerard Kleywegt
Published 7 March 2012
-
![]()
From Curiosity to Cure
Marc Kvansakul
Published 13 December 2011
![]()
The Lure of the Sandbox
Paul Emsley and Coot
Published 15 October 2011
-
![]()
Springsteen, Tolkien, Protein
Alwyn Jones and Frodo
Published 17 June 2011
![]()
Structures Solved Simply
Paul Adams and Tom Terwilliger on Phenix
Published 2 June 2011
-
![]()
Playing the Odds
Randy Read and Phaser
Published 19 May 2011
![]()
Escape from the Darkroom
Wolfgang Kabsch and XDS
Published 19 May 2011
-
![]()
Better, Faster, Stronger, More
Victor Lamzin and ARP/wARP
Published 17 May 2011
![]()
Crystallography for Kids
Lynne Howell
Published 17 May 2011





































































































































