Resolution solutions
Willy Wriggers
Old Dominion University, Norfolk, Virginia, USA
Published February 25, 2022
A childhood gift of a microscope appears in the origin stories of many scientists, but usually it’s the seen object, with its startling new clarity, that inspires life-long curiosity. For Willy Wriggers, the fascinating aspect was the microscope itself.
A preference for the tools of the trade landed him on the front lines of cryo-electron microscopy (cryo-EM) in the technique’s early days. For more than two decades, structural biologists have been deploying his software to make sense of intermediate-resolution data they generate in their laboratories.
Wriggers, a computational biophysicist at Old Dominion University in Norfolk, Virginia, may be best known for a set of software tools called Situs. The software helps researchers construct atomic resolution models from low-resolution single-particle cryo-electron microscopy (cryo-EM), small x-ray scattering and electron tomography data. The software is available at SBGrid and http://situs.biomachina.org/ .
In the early days of cryo-EM, rigid-body and flexible fitting of atomic resolution fragments was the only option to interpret the relatively low-resolution data. These days, direct detection cameras and other technical advances allow atomic resolution more routinely. But “since the field grew exponentially in importance, fitting and detection of secondary structure features in intermediate resolution maps are still quite important,” Wriggers says.
For example, many cryo-EM researchers still make a lower resolution first pass through their molecular data, he says, or there may be lower resolutions portions lingering in a high-resolution structure.
And then there is the new frontier of electron tomography, with the ultimate goal to capture the three-dimensional structure of molecular complexes in their native environments, such as imaging an entire cell, with its organelles and cytoskeleton.
“We can develop new techniques for that specific purpose,” he says. “And that I find intellectually challenging and rewarding. Instead of imaging multiple particles, you are now averaging a single cell from multiple directions, but at a very low dose, which introduces a lot of noise in your images.” His lab’s work currently focuses on tracing actin filaments but next on the list is membranes, ribosomes and other types of organelles.
Wriggers grew up near Ingolstadt, Germany, the son of a window manufacturer (dad) and real estate agent (mom). The city in Bavaria is the site of the headquarters of the German automobile manufacturer Audi. Its turbocharged rally cars were an obsession among local youth in the 1980s and attracted a whole generation to engineering and motorsports, Wriggers says.
His uncle gave him his first microscope. After school, he prowled backyards in his neighborhood looking for samples to examine. He told his parents he wanted to be a scientist. He relished the starring role of scientists in classic 1950s science fiction movie reruns he watched late at night on an old black and white television in his room. He was riveted by the dozens of missions launched in NASA’s space shuttle program and by the Voyager planetary probes.
At age 12, he had graduated from microscopes to telescopes and joined the Ingolstadt astronomy club, whose young members had just built an observatory on the high school roof. There he tinkered with solar observing after school and deep-sky imaging at night. By age 16, he was editing the club’s magazine “Rosa Ursina” and giving public stargazing tours and education presentations in cosmology—a preview of his future life as a professor, he jokes.
Following his interests in astronomy he studied physics at nearby University of Regensburg in 1989, where he was exposed to computers for the first time. “That changed everything for me, because I was just super fascinated at the time with fractals, nonlinear dynamics and chaos theory and their application to the stability of planetary orbits.”

In 1992, an academic exchange stipend gave him the chance to pursue a physics PhD at University of Illinois Urbana-Champaign, which had one of the leading centers in nonlinear dynamics. It was a heady time to be on that campus. One floor up from his lab at the Beckman Institute was the National Center of Supercomputing Applications, where two programmers released the first browser (Mosaic) for the nascent World Wide Web.
In the lab of Klaus Schulten, Wriggers started working with neural networks and machine learning and then transitioned to the lab’s new exclusive focus on more applied molecular dynamics simulations of motor proteins. At scientific meetings, he listened to researchers talk about how they fit atomic structures onto electron microscopy maps by hand. He saw the potential of combining the neural network algorithms with the simulations.
In 1997, he moved to San Diego to a joint postdoc with Andrew McCammon at University of California, San Diego (UCSD) and with Ronald Milligan at The Scripps Research Institute. Milligan studied actin-myosin motor proteins and was a pioneer at fitting atomic structures by hand. Wriggers dove into computational methods to dock smaller x-ray crystal structures on the large and fuzzy volume maps from EM.
The next year, Gina Sosinsky, a UCSD colleague, advised Wriggers to pull a paper together in the next month for a special issue of the journal of Structural Biology. He rushed his collection of software scripts onto the Internet to be freely downloaded by anyone. It took off. One year later, he had a faculty position at Scripps and his first grant.
In 2003, he moved to University of Texas Health Science Center at Houston where he held his first independent position. The collegial atmosphere among the structural biologists in the Houston area reinforced his preference for the tools and the opportunity to bring techniques from other computational fields into modeling.
In 2007, Wriggers moved to New York City to participate in the development of the Anton supercomputer at D. E. Shaw Research. His work there culminated in the first millisecond length molecular dynamics simulation in 2010. This was a revolutionary breakthrough at the time. “Physics-based force fields successfully folded proteins in the computer,” he says, “although the long simulations also showed that force fields do not capture all the relevant interactions, so deep learning approaches such as AlphaFold have more predictive power nowadays.”
Meanwhile, the cryo-EM field was on the cusp of its own revolution in 2014, when direct detection cameras enabled the direct solving of atomic structures, and it became the dominant method in structural biology.
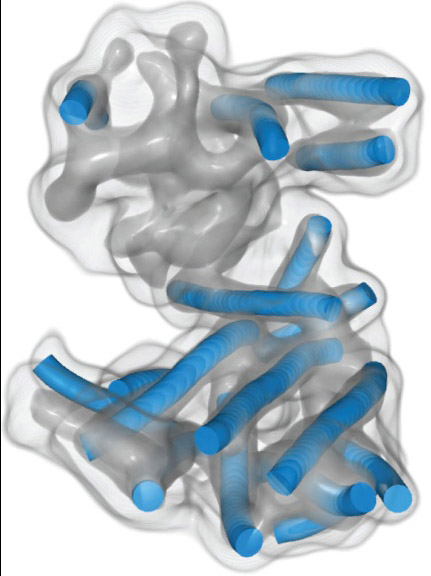
Throughout his stint in industry, Wriggers kept his cryo-EM research alive, initially with a grant-funded team in Houston and virtually after 2012, while searching for a new home for his NIH funded project. The volunteer virtual team included programmer Manuel Wahle (now at Montefiore Medical Center in New York), postdoc Zbigniew Starosolski (now research faculty at Texas Children’s Hospital in Houston) and Ph.D. student Mirabela Rusu (now a professor at Stanford University, she pioneered the detection of alpha-helices in intermediate resolutions cryo-EM maps).
In 2014, Wriggers returned to academia and joined the faculty at Old Dominion University as an endowed chair of mechanical and aerospace engineering, where he was able to renew his long-standing NIH grant in cryo-EM computation in 2015 and 2020.
“Structural biology is the major application domain for me, but it's not the driving motivation,” he says. “The driving motivation dating back to my physics years has always been numerical computing and also the maths behind it.”
In the new engineering environment, he also has found computational inspiration in new and familiar domains, such as space research and race car performance. For example, for a university-sponsored race team, he developed a strategy to improve lap times based on car sensor data and applied multivariate statistics on that sensor data to improve the engineering of the cars. He also worked on a numerical method for mapping magnetic fields in satellites.
In structural biology, Wriggers started a new project at Old Dominion with a computational colleague, Jing He, to develop new ways to blend low- and medium-resolution data for multi-scale modeling. He collaborates extensively with structural biology labs at other universities over a wide range of biological systems. He has retained a special interest in actin and myosin filaments.

In one collaboration, Wriggers and He held weekly Skype sessions every Friday afternoon with Manfred Auer of Lawrence Berkeley National Laboratory. Auer uses electron tomography to study inner ear hair cells, which are central to hearing and balance. The bundles of hair-like projections called stereocilia that sense sound waves are filled with actin filaments that taper off at the tips. Wriggers and He are developing and validating a new approach to the tedious and time-consuming task of tracing actin filaments by hand in whole cell imaging.
Wriggers lives in the Chic’s Beach community of Virginia Beach with his wife Hilary (a biomedical laboratory manager) and two sons. The family learned various instruments at the nearby School of Rock, and you can find Wriggers and his sons performing at open mics and on the local blues jam circuit at venues like Froggies or Jerry’s Indian River.
Carol Cruzan Morton





































































































































