Frame by frame
Nikolaus Grigorieff
HHMI’s Janelia Research Campus
Published January 28, 2019
Rejection is an unavoidable part of science, whether it’s funding for research or manuscripts submitted for publication. But few people showcase their setbacks like Nikolaus “Niko” Grigorieff, whose lab web site features a separate “rejection” section.
As a methods developer in structural biology, Grigorieff may be best known for pioneering a technique known as motion correction in cryo-electron microscopy (cryo-EM). But the numbers tell a different story.
All together, his papers have been cited about 9,500 times, excluding self-citations. Compare that to the 45,000 downloads of rejection letters his web site logged as of January 2019 for the 20 papers (so far) that were vetoed by one or more journals before being published.
“People love reading these rejections,” says Grigorieff, who started posting them in 2010 after lab discussions of frustration with the peer review process.
One notable rejection involves one of the group’s pivotal motion correction studies. It demonstrated how taking movies and analyzing them frame by frame can restore atomic detail to blurred molecular images. The rejection letters from Nature Methods and PNAS are posted with the PDF of the 2012 paper in Structure. “After this paper was published, everybody in the cryo-EM field who has a movie-capable camera switched to movies,” he comments on the entry.
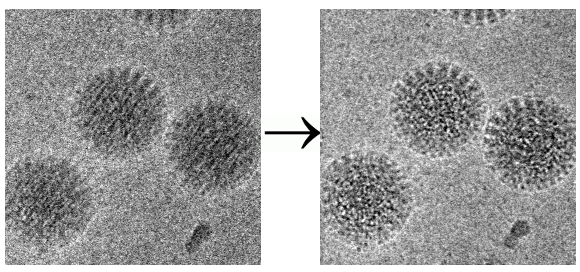
Other authors are welcome to contribute their rejection letters as soon as a paper is accepted elsewhere, but so far only one other research lab has volunteered a sample.
Grigorieff grew up in Berlin, Germany. He didn’t like to hang out with people, preferring to fiddle with electronics and computers at home. His fix-it skills made him popular with friends and family, at least most of the time. After his mom turned off the electricity because he was playing his music too loud, he built a generator to power his stereo independently.
He even wrote his own computer games, including a strategic game that beat him a few times. These days, online gaming helps him maintain family ties. Since he moved away from Germany after his undergraduate degree, he has a Skype chat with his brother almost every Sunday, and then they play an online game together.
Grigorieff picked up his EM skills during his physics PhD training at the University of Bristol in the UK. For postdoctoral work, he moved to the Cambridge lab of Richard Henderson, where he encountered proteins and the related cryo-EM motion problem.
In materials science, “you switch on the electron beam, the sample starts moving, and you wait until it stops to take a picture. There, you can get high resolution without movies,” Grigorieff says. “But with a protein or a cell, it’s getting more and more damaged while you wait. By the time it stops moving, there's nothing left. You have to switch on the beam and take a picture immediately when the sample is still intact. But you had to accept that it was always a little blurred because of this movement.”
At first, Grigorieff helped develop the two-dimensional electron crystallography in the Henderson lab’s push to higher resolution. Soon, his interest was captured by single-particle cryo-EM, which could not then be used to build a three-dimensional atomic model. Two main research groups had developed software (SPIDER from Joachim Frank and IMAGIC from Marin van Heel). Grigorieff found a few processing steps that could be done more efficiently, so he began to develop what would become Frealign, a computer program to refine single-particle structures and correct for image distortions commonly found in electron microscope images.
After five years at MRC Laboratory of Molecular Biology, Grigorieff heard about a cryo-EM opening at Brandeis University near Boston, Mass. He was ready for a new adventure, and a move to the United States would put him closer to his then girlfriend in Arizona. He joined the Brandeis faculty in 1999 and one year later secured long-term funding as an investigator of the Howard Hughes Medical Institute.
“As a Hughes investigator, you can have crazy ideas,” he says. “We could try out things that otherwise would have been harder or impossible. It’s an enormous freedom.”
At Brandeis, he continued to develop Frealign. He also developed other single-particle EM tools and applied them in collaborations with other structural biology groups. One milestone was CTFFIND, an important step in achieving accurate high resolution with cryo-EM that became the industry standard. These tools have been refined and integrated with many others in a complete user-friendly software package called cisTEM, published in eLife March 2018, that covers the pipeline from imaging to 3D reconstruction for single particle EM.
At home, Grigorieff can sit for hours in the backyard with a telescope and a beer, looking at the quiet night sky and photographing celestial objects. Like samples caught in the cryo-EM beam, objects move across the sky. To take a photograph, a computer takes the motion into account, guiding the telescope and camera to track the object over the hours needed to collect an image.

For proteins shaken up by the electron beam, several scientists had proposed taking movies instead of pictures and aligning the movie frames to undo the blurring that normally occurs.
Suddenly, in 2011, there was a new experimental technology that could achieve this. Called a direct detector, the camera directly and quickly recorded electrons reflected off the protein sample. Three companies were developing direct detectors in collaboration with research groups, one of which was located in San Diego. Time for a sabbatical. In Boston, Grigorieff had met and married the niece of the immigration lawyer handling his green card, the children’s book author Maria Gianferrari. “We packed up the whole family, dog included, and drove from Massachusetts to San Diego for 13 months,” he says.
The sabbatical project resulted in the first report of this kind of movie collection and image correction in 2012 in the Journal of Structural Biology. First author Axel Brilot, a graduate student with Grigorieff at Brandeis, was honored with a "Paper of the Year" award in 2015. And in the scientific background for the 2017 Nobel Prize in Chemistry for development of cryo-EM awarded to Henderson, Frank, and Jacque Dubochet, the 2012 paper was cited as one of the recent advances that built on the earlier groundbreaking work by the three scientists. (The follow-up paper that demonstrated how well the technique works was rejected twice before being published.)
In 2012, Grigorieff returned to Brandeis. He realized cryo-EM was about to change, with expensive new microscopes and detectors that needed a critical mass of scientists for support. He could have tried to forge a bigger alliance in the Boston area, but he had a connection with HHMI’s Janelia Research Campus. He had helped install the first big cryo-EM instrument and had nearly accepted an offer to be a founding investigator there a decade earlier. It made more sense to him to take advantage of a new shared facility for investigators at Janelia in Virginia, where he could also devote 100% of his time to research.
With his HHMI funding recently renewed, Grigorieff will move the family back to New England in 2020, when his daughter finishes high school. This time, he will work at University of Massachusetts Medical School in Worcester. He is contemplating his next big scientific direction.
He may follow up on a feasibility study to image proteins inside cells proposed with collaborators Peter Rickgauer and Winfried Denk of the Max Planck Institute of Neurobiology in Germany. They outlined their approach in a May 2017 paper in eLife. The paper proposed a way of studying single proteins in the crowded environment of the cell, using single particle cryo-EM to catch them in interactions with other molecules.
Structural biologists purify proteins they want to study, making inferences about their function and activity from the shape and chemistry of the atomic structure. Cell biologists wonder how scientists can be sure proteins work that way inside living cells, Grigorieff says. “There’s a lot you can learn from purified protein, but it’s a valid point.”
There are many caveats and obstacles to overcome. The structure of the proteins would need to be known to find them inside the cell, they would have to be on the bigger side, and the cells themselves would be frozen, stopping all the interesting movements. Yet, it should be possible to determine what is next to what and how molecules interact in these cells to bring them to life, says Grigorieff. “It’s early days.”
--Carol Cruzan Morton
Other tales
-
![]()
The Stories Antibodies Tell
Jean-Philippe Julien
Published 27 January 2026
![]()
Reshaping Membranes
Melanie Ohi
Published 23 November 2025
-
![]()
Probing Microbes
Gira Bhabha
Published 30 September 2025
![]()
Drawn to the Light
Emina Stojković
Published 30 July 2025
-
![]()
The Final Phase
George Phillips
Published 31 May 2025
![]()
Mind and Muscle
Ryan Hibbs
Published 28 March 2025
-
![]()
The Shapes of Energy
Luke Chao
Published 12 December 2024
![]()
Predicting Proteins
Jens Meiler
Published 25 November 2024
-
![]()
Death Metal
Steven Damo
Published 28 April 2024
![]()
Context Matters
Bing Chen
Published 30 January 2024
-
![]()
The Crystal Whisperer
Sarah Bowman
Published 29 November 2023
![]()
Data in Motion
Nozomi Ando
Published 29 September 2023
-
![]()
The Monstrous Maw
André Hoelz
Published 28 June 2023
![]()
Second Takes
Andrea Thorn
Published 28 February 2023
-
![]()
Radical reactions
Yvain Nicolet
Published 31 January 2023
![]()
Floppy Physics
Eva Nogales
Published 30 November 2022
-
![]()
Structure of Equity
Jamaine Davis
Published 28 September 2022
![]()
Life and Death of a Cell
Evris Gavathiotis
Published 28 July 2022
-
![]()
Follow the glow
Kurt Krause
Published 29 April 2022
![]()
Resolution solutions
Willy Wriggers
Published 25 February 2022
-
![]()
Of enzymes and membranes
Ming Zhou
Published 28 October 2021
![]()
Step-by-step
Gabrielle Rudenko
Published 26 September 2021
-
![]()
Moving muscle
Montserrat Samso
Published 26 July 2021
![]()
Particle catcher
Stefan Raunser
Published 28 June 2021
-
![]()
Designer drugs
Ho Leung Ng
Published 25 February 2021
![]()
Right place, right time
Ernesto Fuentes
Published 29 January 2021
-
![]()
Shape-shifting secrets of membranes
James Hurley
Published 27 November 2020
![]()
Enzymatic action
Cynthia Wolberger
Published 28 September 2020
-
![]()
Rules of motion
Priyamvada Acharya
Published 31 July 2020
![]()
Cosmic Squared
Michael Cianfrocco
Published 27 June 2020
-
![]()
Kaps are Cool
Yuh Min Chook
Published 28 April 2020
![]()
Spiraling into focus
Carsten Sachse
Published 29 March 2020
-
![]()
Seeing cilia
Alan Brown
Published 27 February 2020
![]()
For the Love of EM
Guy Schoehn
Published 27 January 2020
-
![]()
Protein Puddles
Michael Rosen
Published 16 December 2019
![]()
Changing channels
Daniel Minor Jr.
Published 27 September 2019
-
![]()
Listening Tips
Marcos Sotomayor
Published 30 July 2019
![]()
Beyond Cool
Published 31 May 2019
-
![]()
Hao Wu
A Higher Order
Published 30 May 2019
![]()
Aye Aye Captain
Alexandre Bonvin
Published 29 April 2019
-
![]()
The PARP Family Family
John Pascal
Published 28 February 2019
![]()
Frame by frame
Nikolaus Grigorieff
Published 28 January 2019
-
![]()
Predicting Success
Bil Clemons
Published 18 December 2018
![]()
Curiouser and Curiouser
Ramaswamy Subramanian
Published 27 November 2018
-
![]()
Rely on This
Sjors Scheres
Published 26 October 2018
![]()
Proteins out of bounds
Gerhard Wagner
Published 27 September 2018
-
![]()
Hiding in plain sight
Gaya Amarasinghe
Published 27 July 2018
![]()
Jumping Genes
Orsolya Barabas
Published 27 June 2018
-
![]()
Data Whisperer
Karolin Luger
Published 30 May 2018
![]()
Flipping the Switch
Jacqueline Cherfils
Published 27 April 2018
-
![]()
Tooling Around
Andrew Kruse
Published 29 March 2018
![]()
Comings and Goings
Tom Rapoport, Ph.D.
Published 23 February 2018
-
![]()
Transcriptional Rhythm
Seth Darst
Published 27 January 2018
![]()
The Language of Gene Regulation
Daniel Panne
Published 21 November 2017
-
![]()
Not Your Average Protein
James Fraser
Published 23 October 2017
![]()
Message Received
Sebastien Granier
Published 24 August 2017
-
![]()
Resistance is Futile
Celia Schiffer
Published 28 July 2017
![]()
Twist of Fate
Leemor Joshua-Tor
Published 28 June 2017
-
![]()
Drug Designer
John Buolamwini
Published 30 May 2017
![]()
Mathematically Minded
James Holton
Published 28 April 2017
-
![]()
Garbage Out
Kay Diederichs
Published 30 March 2017
![]()
Fixer Upper
Brandt Eichman
Published 27 February 2017
-
![]()
Mobilizers
Phoebe Rice
Published 31 January 2017
![]()
Escape Artist
Katya Heldwein
Published 19 December 2016
-
![]()
Nature’s Confectioner
Jochen Zimmer
Published 29 November 2016
![]()
State of Fusion
Jason McLellan
Published 27 October 2016
-
![]()
Here Be Dragons
Brian Fox
Published 28 September 2016
![]()
SBGrid Assumes Ownership of PyMOLWiki
Published 15 September 2016
-
![]()
Pharm Team
Oleg Tsodikov
Published 24 August 2016
![]()
Spiro-Gyra
Alejandro Buschiazzo
Published 27 July 2016
-
![]()
Turning the DIALS
Nicholas Sauter
Published 29 June 2016
![]()
Pipeline Dreams
Bridget Carragher and Clint Potter
Published 26 April 2016
-
![]()
U-Store-It
The SBGrid Data Bank provides an affordable and sustainable way to preserve and share structural biology data
Published 28 March 2016
![]()
Big Questions, Big Answers
Jennifer Doudna
Published 22 February 2016
-
![]()
Not a Structural Biologist
Enrico Di Cera
Published 17 December 2015
![]()
Divide and Conquer
Kevin Corbett
Published 19 November 2015
-
![]()
Computing Cellular Clockworks
Klaus Schulten
Published 23 October 2015
![]()
Trans-Plant
Gang Dong
Published 26 September 2015
-
![]()
Keep on Moving
James Berger
Published 23 August 2015
![]()
Totally Tubular
Antonina Roll-Mecak
Published 27 July 2015
-
![]()
From Disorder, Function
Julie Forman-Kay
Published 29 June 2015
![]()
Into Alignment
Geoff Barton
Published 27 May 2015
-
![]()
Two Labs, Many Methods
Michael Sattler
Published 28 April 2015
![]()
Picture This
Georgios Skiniotis
Published 20 March 2015
-
![]()
Intron Intrigue
Navtej Toor
Published 20 February 2015
![]()
Cut and Paste
Martin Jinek
Published 28 January 2015
-
![]()
Basics and Beyond
Qing Fan
Published 18 December 2014
![]()
Bloodletting and Other Studies
Pedro José Barbosa Pereira
Published 25 November 2014
-
![]()
Wire Models, Wired
A brief history of UCSF Chimera
Published 29 October 2014
![]()
In Search of…New Drugs
Doug Daniels
Published 30 September 2014
-
![]()
An Affinity for Affinity…and Corals
John C. Williams
Published 29 August 2014
![]()
Pete Meyer, Ph.D.
Research Computing Specialist
Published 22 August 2014
-
![]()
Justin O'Connor
Sr. System Administrator
Published 20 August 2014
![]()
Carol Herre
Software Release Engineer
Published 15 August 2014
-
![]()
Elizabeth Dougherty
Science Writer
Published 13 August 2014
![]()
Andrew Morin, Ph.D.
Policy Research Fellow
Published 11 August 2014
-
![]()
Jason Key, Ph.D.
Associate Director of Technology and Innovation
Published 8 August 2014
![]()
Piotr Sliz, Ph.D.
Principal Investigator, SBGrid
Published 1 August 2014
-
![]()
New Kid on the Block
James Chen
Published 29 July 2014
![]()
Membrane Master
Tamir Gonen
Published 30 June 2014
-
![]()
The Natural Bridge
Piotr Sliz
Published 13 June 2014
![]()
Surprise, Surprise
Catherine Drennan
Published 26 April 2014
-
![]()
Gone Viral
Olve Peersen
Published 20 March 2014
![]()
All Who Wander Are Not Lost
Frank Delaglio
Published 24 February 2014
-
![]()
The Raw and the Cooked
Graeme Winter
Published 24 January 2014
![]()
Vacc-elerator
Peter Kwong
Published 17 December 2013
-
![]()
Structural Storyteller
Karin Reinisch
Published 15 November 2013
![]()
The Fixer
Jane Richardson
Published 28 October 2013
-
![]()
Inside the Box
Mishtu Dey
Published 17 September 2013
![]()
Sensing a Change
Brian Crane
Published 16 August 2013
-
![]()
Towards Personalized Oncology
Mark Lemmon
Published 16 July 2013
![]()
Brush with Fame
Yizhi Jane Tao
Published 14 June 2013
-
![]()
Toxic Avenger
Borden Lacy
Published 21 May 2013
![]()
Pushing the Boundaries
Stephen Harrison
Published 22 April 2013
-
![]()
Strength in Numbers
Joseph Ho
Published 18 March 2013
![]()
One Lab, Many Methods
Wesley Sundquist
Published 12 February 2013
-
![]()
Unplanned Pioneer
Tim Stevens
Published 15 January 2013
![]()
From Actin to Action
Emil Pai
Published 11 January 2013
-
![]()
Unstructured
A Brief History of CCP4
Published 12 December 2012
![]()
Stop, Collaborate and Listen
Eleanor Dodson
Published 5 November 2012
-
![]()
X-PLORer
Axel Brunger
Published 1 October 2012
![]()
Share the Wealth
Zbyszek Otwinowski
Published 22 August 2012
-
![]()
Unraveling RNA
Anna Pyle
Published 18 July 2012
![]()
Sharper Image
Pawel Penczek and SPARX
Published 4 June 2012
-
![]()
Creative Copy Cat
Pamela Bjorkman
Published 25 April 2012
![]()
Charm and Diplomacy
Gerard Kleywegt
Published 7 March 2012
-
![]()
From Curiosity to Cure
Marc Kvansakul
Published 13 December 2011
![]()
The Lure of the Sandbox
Paul Emsley and Coot
Published 15 October 2011
-
![]()
Springsteen, Tolkien, Protein
Alwyn Jones and Frodo
Published 17 June 2011
![]()
Structures Solved Simply
Paul Adams and Tom Terwilliger on Phenix
Published 2 June 2011
-
![]()
Playing the Odds
Randy Read and Phaser
Published 19 May 2011
![]()
Escape from the Darkroom
Wolfgang Kabsch and XDS
Published 19 May 2011
-
![]()
Better, Faster, Stronger, More
Victor Lamzin and ARP/wARP
Published 17 May 2011
![]()
Crystallography for Kids
Lynne Howell
Published 17 May 2011





































































































































