Comings and Goings
Tom Rapoport, Ph.D.
HHMI, Harvard Medical School
Published February 23, 2018
Cell biologist Tom Rapoport may be best known for studies of how proteins get in and out of a convoluted compartment inside cells called the endoplasmic reticulum (ER). But his personal backstory rivals his scientific achievements as a Howard Hughes Medical Institute Investigator at Harvard Medical School (HMS). His life has been intimately shaped by major political persecutions and social upheavals of 20th century Europe and America. Here’s the short version:
“My father was born in Russia into a Jewish family that emigrated during the Russian revolution to Vienna,” Rapoport wrote in a 2010 essay. “[In] the 1920s, [he] became a member of the Socialist party and later of the Communist party. He studied medicine and chemistry. While he was on a fellowship in Cincinnati, Ohio, the Nazis took over Austria and he could not return. In Cincinnati, he met my half-Jewish mother, who had emigrated from Nazi Germany and restarted her career as a pediatrician. I was born in Cincinnati, but 3 years later, in 1950, my parents became targets of the anticommunist campaign of Joseph McCarthy and returned to Europe, initially for a year in Austria. Because my dad was blacklisted, he could not find a job in Austria and we moved to East Germany, where he joined the faculty of Humboldt University in East Berlin. I was 4 years old when we arrived, and I stayed in Berlin until age 48.” In 1995, Tom and eight other members of his lab boarded a plane to Boston, where he has happily settled. He returns to hike in the Alps every year with at least one of his children.
Science begins at home
Rapoport credits his parents for his interest in science. He recalls stimulating discussions at the dinner table. His success in the Math Olympiads enabled him to go to a special high school for math and science. He developed a love for chemistry, thanks in part to Linus Pauling’s General Chemistry.

Rapoport attended Humboldt University on a special research program that combined undergraduate work with a PhD. East Germany’s PhD program also allowed two doctoral students to team up. Halfway through the seven-year program, he switched to biochemistry, putting him in an institute headed by his father. Rapoport ultimately earned two pairwise postgraduate degrees, but not before accidentally leaving a faucet on, flooding three floors of the institute, including his father’s office. His father made him renovate all of the damaged spaces.
The main publication in 1974 from his second doctorate remains his most-cited paper. The thesis, in partnership with Reinhart Heinrich, “described in quantitative terms the importance of an enzyme for the overall flux through a metabolic pathway and for the regulation of metabolite concentrations,” Rapoport recalls. The theory, now known as “metabolic control analysis (MCA),” was independently developed by another team at the same time and is lauded as an early contribution to the field of systems biology.
Moving to another institute in East Berlin, Rapoport became interested in protein translocation during a project to clone insulin mRNA. Fish, he learned, had larger Islets of Langerhans than mammals. On fish days, lab members lined up around a table to harvest the fresh carp parts they needed. (The rest of the fish was sold cheaply to grateful people lined up in front of the lab.) As part of that work, the group determined the first protein and gene sequences in East Germany.
Membrane ins and outs
Rapoport now wondered how a polypeptide’s zip-code like signal sequence is recognized by the membrane and how the polypeptide then moves through the membrane. With his students, Rapoport reported the first evidence that Sec61p forms the essential protein-conducting channel through the ER membrane for newly made proteins. Rapoport realized the similarities with the related bacterial SecY channel and proposed a unifying concept of protein translocation in 1994.
Science was difficult in East Germany, because of the isolation and lack of resources. When Rapoport made a rare trip to the United States, the FBI called each university host after he left to verify he was a “real scientist.” After the Berlin wall came down in 1989 and Germany was reunified in 1990, Rapoport suddenly found his grant applications funded in full. Yet, even the best and brightest East German scientists faced residual prejudices from their West German colleagues, who were now largely in charge at newly merged universities. Rapoport was denied a professorship at the new institution because of his past engagement in communist East Germany.
At Harvard, Rapoport continued to advance the field of protein translocation. In three main projects, his group has been working out how proteins cross into the ER membrane, how misfolded ER proteins are ejected back (“retrotranslocation”) into the cell cytosol for destruction (“ER-associated protein degradation, or ERAD”), and how the ER gets its shape. The ER is crucial for producing certain proteins and most of the lipids needed by other cell organelles. It occupies 10 percent of a cell’s volume and accounts for nearly half the membrane found in a typical animal cell.
While he insists much remains to be learned about ERAD in particular, Rapoport recently added a fourth project, the mechanism of protein import into peroxisomes, to attract students and postdoctoral fellows who want to make an impact on a less-studied biological problem. Peroxisomes are involved in energy metabolism and detoxification, and their malfunction results in serious human diseases.
In a nascent project, Rapoport also recently became interested in lung surfactants after glimpsing “amazing EM pictures” of the onion-like lipid bilayers in aveolar cells. Premature babies need surfactant spray, because their lung cells cannot yet generate surfactant.
“Every project has something to do with membranes,” Rapoport says. “My lab is known for using purified proteins and reconstituting a process with purified proteins. We start out with some murky cell biology problem – how proteins get across the membrane, how the shape of the membrane determines its function— and eventually understand it at molecular level.”
Shape of things to come
To learn these mechanistic details, the lab solves a lot of structures, often in collaboration with others. One highlight is the 2004 X-ray structure of an archaebacterial homolog of the Sec61p complex (with Stephen Harrison, a fellow Howard Hughes investigator at HMS). In a major result in 2016, they captured a structure of the active channel with a translocating polypeptide chain being pushed through.
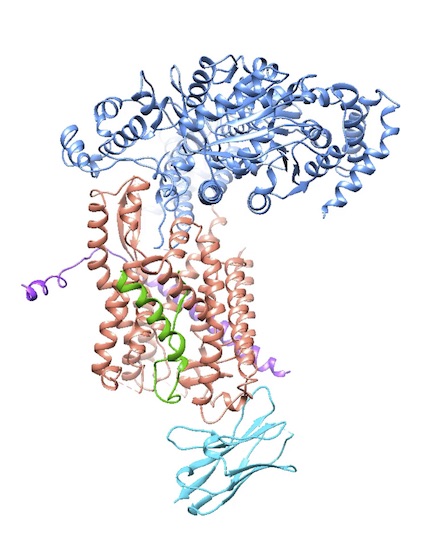
For proteins that do not fold correctly in the ER and retrotranslocate back to the cytosol, Rapoport’s group is working on a comprehensive picture of the ERAD pathway in yeast. In 2017, electron microscopy (EM) gave them a first glimpse of how the crucial Hrd1 channel allows proteins to cross the membrane. Ideally, they will catch Cdc48 ATPase, with its ring-like structure, pulling a misfolded protein back through the membrane.
The lab has EM collaborations with Tom Walz at Rockefeller University, Maofu Liao at HMS, and Christopher Akey at Boston University and has recently built independent EM capabilities as well. In June 2016, Rapoport decided he wanted to do more lab work, starting with crystallizing molecules too small for EM analysis. He found it a breeze: One-third of his trays grew crystals. His philosophy is to tackle the biological problem first, being flexible about learning whatever method is required to answer the question.
Rapoport, who turned 70 in 2017, anticipates a thriving lab for at least another decade or so. In this, too, there is family precedence. After all, his mother finally passed her PhD oral exam at age 102 in 2015, 77 years after her thesis on diphtheria was labeled with a yellow stripe, as Jews would soon be forced to wear yellow stars on their sleeves, and her oral exam canceled.
Looking ahead to where cell biology is heading, Rapoport advises young people to go into organ-specific cell biology, rather than trying to work on the fundamental processes of his generation. “Every cell type in a higher organism has a particular biology,” he says. “Major discoveries will be made and will be more medically relevant than in the past.”
-Carol Curzan Morton
Other tales
-
![]()
The Stories Antibodies Tell
Jean-Philippe Julien
Published 27 January 2026
![]()
Reshaping Membranes
Melanie Ohi
Published 23 November 2025
-
![]()
Probing Microbes
Gira Bhabha
Published 30 September 2025
![]()
Drawn to the Light
Emina Stojković
Published 30 July 2025
-
![]()
The Final Phase
George Phillips
Published 31 May 2025
![]()
Mind and Muscle
Ryan Hibbs
Published 28 March 2025
-
![]()
The Shapes of Energy
Luke Chao
Published 12 December 2024
![]()
Predicting Proteins
Jens Meiler
Published 25 November 2024
-
![]()
Death Metal
Steven Damo
Published 28 April 2024
![]()
Context Matters
Bing Chen
Published 30 January 2024
-
![]()
The Crystal Whisperer
Sarah Bowman
Published 29 November 2023
![]()
Data in Motion
Nozomi Ando
Published 29 September 2023
-
![]()
The Monstrous Maw
André Hoelz
Published 28 June 2023
![]()
Second Takes
Andrea Thorn
Published 28 February 2023
-
![]()
Radical reactions
Yvain Nicolet
Published 31 January 2023
![]()
Floppy Physics
Eva Nogales
Published 30 November 2022
-
![]()
Structure of Equity
Jamaine Davis
Published 28 September 2022
![]()
Life and Death of a Cell
Evris Gavathiotis
Published 28 July 2022
-
![]()
Follow the glow
Kurt Krause
Published 29 April 2022
![]()
Resolution solutions
Willy Wriggers
Published 25 February 2022
-
![]()
Of enzymes and membranes
Ming Zhou
Published 28 October 2021
![]()
Step-by-step
Gabrielle Rudenko
Published 26 September 2021
-
![]()
Moving muscle
Montserrat Samso
Published 26 July 2021
![]()
Particle catcher
Stefan Raunser
Published 28 June 2021
-
![]()
Designer drugs
Ho Leung Ng
Published 25 February 2021
![]()
Right place, right time
Ernesto Fuentes
Published 29 January 2021
-
![]()
Shape-shifting secrets of membranes
James Hurley
Published 27 November 2020
![]()
Enzymatic action
Cynthia Wolberger
Published 28 September 2020
-
![]()
Rules of motion
Priyamvada Acharya
Published 31 July 2020
![]()
Cosmic Squared
Michael Cianfrocco
Published 27 June 2020
-
![]()
Kaps are Cool
Yuh Min Chook
Published 28 April 2020
![]()
Spiraling into focus
Carsten Sachse
Published 29 March 2020
-
![]()
Seeing cilia
Alan Brown
Published 27 February 2020
![]()
For the Love of EM
Guy Schoehn
Published 27 January 2020
-
![]()
Protein Puddles
Michael Rosen
Published 16 December 2019
![]()
Changing channels
Daniel Minor Jr.
Published 27 September 2019
-
![]()
Listening Tips
Marcos Sotomayor
Published 30 July 2019
![]()
Beyond Cool
Published 31 May 2019
-
![]()
Hao Wu
A Higher Order
Published 30 May 2019
![]()
Aye Aye Captain
Alexandre Bonvin
Published 29 April 2019
-
![]()
The PARP Family Family
John Pascal
Published 28 February 2019
![]()
Frame by frame
Nikolaus Grigorieff
Published 28 January 2019
-
![]()
Predicting Success
Bil Clemons
Published 18 December 2018
![]()
Curiouser and Curiouser
Ramaswamy Subramanian
Published 27 November 2018
-
![]()
Rely on This
Sjors Scheres
Published 26 October 2018
![]()
Proteins out of bounds
Gerhard Wagner
Published 27 September 2018
-
![]()
Hiding in plain sight
Gaya Amarasinghe
Published 27 July 2018
![]()
Jumping Genes
Orsolya Barabas
Published 27 June 2018
-
![]()
Data Whisperer
Karolin Luger
Published 30 May 2018
![]()
Flipping the Switch
Jacqueline Cherfils
Published 27 April 2018
-
![]()
Tooling Around
Andrew Kruse
Published 29 March 2018
![]()
Comings and Goings
Tom Rapoport, Ph.D.
Published 23 February 2018
-
![]()
Transcriptional Rhythm
Seth Darst
Published 27 January 2018
![]()
The Language of Gene Regulation
Daniel Panne
Published 21 November 2017
-
![]()
Not Your Average Protein
James Fraser
Published 23 October 2017
![]()
Message Received
Sebastien Granier
Published 24 August 2017
-
![]()
Resistance is Futile
Celia Schiffer
Published 28 July 2017
![]()
Twist of Fate
Leemor Joshua-Tor
Published 28 June 2017
-
![]()
Drug Designer
John Buolamwini
Published 30 May 2017
![]()
Mathematically Minded
James Holton
Published 28 April 2017
-
![]()
Garbage Out
Kay Diederichs
Published 30 March 2017
![]()
Fixer Upper
Brandt Eichman
Published 27 February 2017
-
![]()
Mobilizers
Phoebe Rice
Published 31 January 2017
![]()
Escape Artist
Katya Heldwein
Published 19 December 2016
-
![]()
Nature’s Confectioner
Jochen Zimmer
Published 29 November 2016
![]()
State of Fusion
Jason McLellan
Published 27 October 2016
-
![]()
Here Be Dragons
Brian Fox
Published 28 September 2016
![]()
SBGrid Assumes Ownership of PyMOLWiki
Published 15 September 2016
-
![]()
Pharm Team
Oleg Tsodikov
Published 24 August 2016
![]()
Spiro-Gyra
Alejandro Buschiazzo
Published 27 July 2016
-
![]()
Turning the DIALS
Nicholas Sauter
Published 29 June 2016
![]()
Pipeline Dreams
Bridget Carragher and Clint Potter
Published 26 April 2016
-
![]()
U-Store-It
The SBGrid Data Bank provides an affordable and sustainable way to preserve and share structural biology data
Published 28 March 2016
![]()
Big Questions, Big Answers
Jennifer Doudna
Published 22 February 2016
-
![]()
Not a Structural Biologist
Enrico Di Cera
Published 17 December 2015
![]()
Divide and Conquer
Kevin Corbett
Published 19 November 2015
-
![]()
Computing Cellular Clockworks
Klaus Schulten
Published 23 October 2015
![]()
Trans-Plant
Gang Dong
Published 26 September 2015
-
![]()
Keep on Moving
James Berger
Published 23 August 2015
![]()
Totally Tubular
Antonina Roll-Mecak
Published 27 July 2015
-
![]()
From Disorder, Function
Julie Forman-Kay
Published 29 June 2015
![]()
Into Alignment
Geoff Barton
Published 27 May 2015
-
![]()
Two Labs, Many Methods
Michael Sattler
Published 28 April 2015
![]()
Picture This
Georgios Skiniotis
Published 20 March 2015
-
![]()
Intron Intrigue
Navtej Toor
Published 20 February 2015
![]()
Cut and Paste
Martin Jinek
Published 28 January 2015
-
![]()
Basics and Beyond
Qing Fan
Published 18 December 2014
![]()
Bloodletting and Other Studies
Pedro José Barbosa Pereira
Published 25 November 2014
-
![]()
Wire Models, Wired
A brief history of UCSF Chimera
Published 29 October 2014
![]()
In Search of…New Drugs
Doug Daniels
Published 30 September 2014
-
![]()
An Affinity for Affinity…and Corals
John C. Williams
Published 29 August 2014
![]()
Pete Meyer, Ph.D.
Research Computing Specialist
Published 22 August 2014
-
![]()
Justin O'Connor
Sr. System Administrator
Published 20 August 2014
![]()
Carol Herre
Software Release Engineer
Published 15 August 2014
-
![]()
Elizabeth Dougherty
Science Writer
Published 13 August 2014
![]()
Andrew Morin, Ph.D.
Policy Research Fellow
Published 11 August 2014
-
![]()
Jason Key, Ph.D.
Associate Director of Technology and Innovation
Published 8 August 2014
![]()
Piotr Sliz, Ph.D.
Principal Investigator, SBGrid
Published 1 August 2014
-
![]()
New Kid on the Block
James Chen
Published 29 July 2014
![]()
Membrane Master
Tamir Gonen
Published 30 June 2014
-
![]()
The Natural Bridge
Piotr Sliz
Published 13 June 2014
![]()
Surprise, Surprise
Catherine Drennan
Published 26 April 2014
-
![]()
Gone Viral
Olve Peersen
Published 20 March 2014
![]()
All Who Wander Are Not Lost
Frank Delaglio
Published 24 February 2014
-
![]()
The Raw and the Cooked
Graeme Winter
Published 24 January 2014
![]()
Vacc-elerator
Peter Kwong
Published 17 December 2013
-
![]()
Structural Storyteller
Karin Reinisch
Published 15 November 2013
![]()
The Fixer
Jane Richardson
Published 28 October 2013
-
![]()
Inside the Box
Mishtu Dey
Published 17 September 2013
![]()
Sensing a Change
Brian Crane
Published 16 August 2013
-
![]()
Towards Personalized Oncology
Mark Lemmon
Published 16 July 2013
![]()
Brush with Fame
Yizhi Jane Tao
Published 14 June 2013
-
![]()
Toxic Avenger
Borden Lacy
Published 21 May 2013
![]()
Pushing the Boundaries
Stephen Harrison
Published 22 April 2013
-
![]()
Strength in Numbers
Joseph Ho
Published 18 March 2013
![]()
One Lab, Many Methods
Wesley Sundquist
Published 12 February 2013
-
![]()
Unplanned Pioneer
Tim Stevens
Published 15 January 2013
![]()
From Actin to Action
Emil Pai
Published 11 January 2013
-
![]()
Unstructured
A Brief History of CCP4
Published 12 December 2012
![]()
Stop, Collaborate and Listen
Eleanor Dodson
Published 5 November 2012
-
![]()
X-PLORer
Axel Brunger
Published 1 October 2012
![]()
Share the Wealth
Zbyszek Otwinowski
Published 22 August 2012
-
![]()
Unraveling RNA
Anna Pyle
Published 18 July 2012
![]()
Sharper Image
Pawel Penczek and SPARX
Published 4 June 2012
-
![]()
Creative Copy Cat
Pamela Bjorkman
Published 25 April 2012
![]()
Charm and Diplomacy
Gerard Kleywegt
Published 7 March 2012
-
![]()
From Curiosity to Cure
Marc Kvansakul
Published 13 December 2011
![]()
The Lure of the Sandbox
Paul Emsley and Coot
Published 15 October 2011
-
![]()
Springsteen, Tolkien, Protein
Alwyn Jones and Frodo
Published 17 June 2011
![]()
Structures Solved Simply
Paul Adams and Tom Terwilliger on Phenix
Published 2 June 2011
-
![]()
Playing the Odds
Randy Read and Phaser
Published 19 May 2011
![]()
Escape from the Darkroom
Wolfgang Kabsch and XDS
Published 19 May 2011
-
![]()
Better, Faster, Stronger, More
Victor Lamzin and ARP/wARP
Published 17 May 2011
![]()
Crystallography for Kids
Lynne Howell
Published 17 May 2011





































































































































