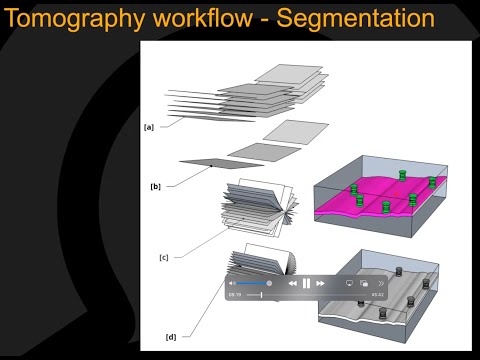
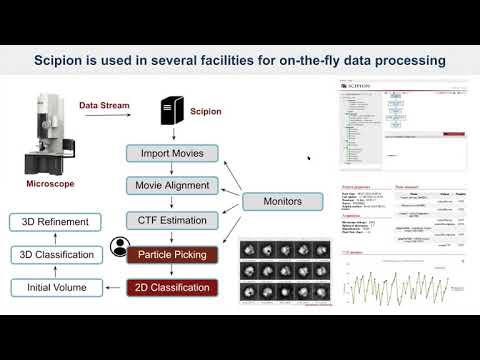
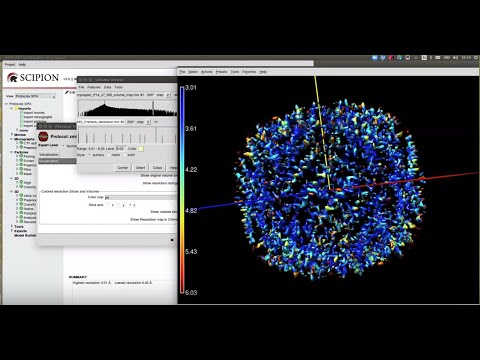
Supported Applications
Scipion
-
Description
an image processing framework to obtain 3D models of macromolecular complexes using 3D EM that allows you to execute workflows combining different software tools, while taking care of formats and conversions.
-
Usage
To list all executables provided by Scipion, run:$ sbgrid-list scipion -
Usage Notes
CryoSPARC plugin is not provided in v3.11.6 (default). But the scipion-em-cryoSPARC plugin can be manually installed in Scipion from SBGrid by the user following the instructions on the original repo, which includes how to link it to an existing CryoSPARC installation: https://github.com/scipion-em/scipion-em-cryosparc2?tab=readme-ov-file#installing-the-plugin
SCIPION uses software that needs Nvidia GPU acceleration. Appropriate hardware and driver are required.
-
Installation
Use the following command to install this title with the CLI client:$ sbgrid-cli install scipionAvailable operating systems: Linux 64 -
Primary Citation*
J. M. de l. Rosa-Trevín, A. Quintana, L. d. Cano, A. Zaldívar, I. Foche, J. Gutiérrez, J. Gómez-Blanco, J. Burguet-Castell, J. Cuenca-Alba, V. Abrishami, J. Vargas, J. Otón, G. Sharov, J. L. Vilas, J. Navas, P. Conesa, M. Kazemi, R. Marabini, C. O. S. Sorzano, and J. M. Carazo. 2016. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. Journal of Structural Biology. 195(1): 93-99.
-
*Full citation information available through
-
-
Citation Note
Scipion is freely available and its source code is 100% open. In exchange, please cite, not only us at Scipion, but ALL of the software that helped you to produce your article: http://scipion.i2pc.es/report_protocols/packages -
Webinars
Topic: ScipionTomo: Towards cryo-electron tomography software integration, reproducibility, and validation.
Presenter: Pablo Conesa Team Leader, Spanish National Research Council
Host: Jason Key
Recorded on October 10, 2023
For more information on Scipion:
https://sbgrid.org/software/titles/scipion
https://github.com/pconesa
-
Keywords
-
Default Versions
Linux 64: 3.11.6 (1.5 GB)
-
Other Versions
Linux 64:
3.7.1 (19.3 GB)
Developers
Jose-Maria Carazo, Jose Miguel de la Rosa Trevin