Supported Applications
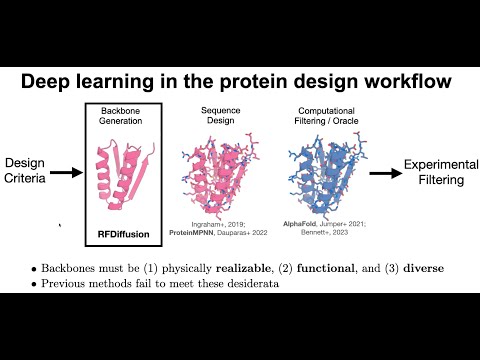
RFdiffusion
-
Description
an open source method for structure generation, with or without conditional information (a motif, target etc). It can perform a whole range of protein design challenges.
-
Usage
To list all executables provided by RFdiffusion, run:$ sbgrid-list rfdiffusion -
Installation
Use the following command to install this title with the CLI client:$ sbgrid-cli install rfdiffusionAvailable operating systems: Linux 64 -
Primary Citation*
J. L. Watson, D. Juergens, N. R. Bennett, B. L. Trippe, J. Yim, H. E. Eisenach, W. Ahern, A. J. Borst, R. J. Ragotte, L. F. Milles, B. I. M. Wicky, N. Hanikel, S. J. Pellock, A. Courbet, W. Sheffler, J. Wang, P. Venkatesh, Sappington I, S. V. Torres, A. Lauko, D. Bortoli V, E. Mathieu, S. Ovchinnikov, R. Barzilay, T. S. Jaakkola, F. DiMaio, M. Baek, and D. Baker. 2023. De novo design of protein structure and function with RFdiffusion. Nature.
-
*Full citation information available through
-
-
Webinars
SBGrid webinars are hosted with partial support from the NIH R25 Continuing Education for Structural Biology Mentors #GM151273, in collaboration with Co-PI Jamaine Davis of Meharry Medical College.
Topic: De novo design of protein structure and function with RFdiffusion.
Presenter: Brian Trippe, Ph.D., Postdoctoral Fellow, Columbia University.
Host: Pete Meyer
Recorded on April 9, 2024
For more information on RFdiffusion:
https://sbgrid.org/software/titles/rfdiffusion
https://github.com/RosettaCommons/RFdiffusion
-
Keywords
Machine Learning, Protein-Protein Interaction Prediction, Protein Structure Analysis
-
Default Versions
Linux 64: 1.1.0 (9.2 GB)
-
Other Versions
Linux 64:
20250912 (11.9 GB)
Developers
David Baker, Joe Watson, Brian Trippe