Supported Applications
DEEPMAINMAST
-
Description
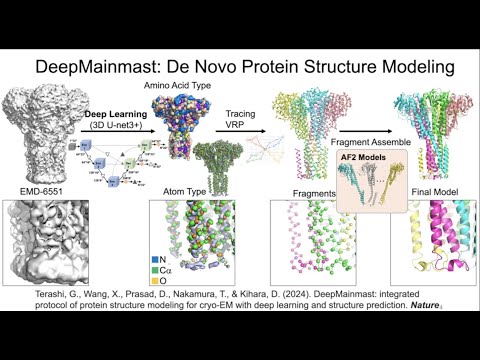
a de novo modeling protocol to build an entire protein 3D model directly from near-atomic (up to about 5 Angstrom) resolution EM map.
-
Usage
To list all executables provided by DEEPMAINMAST, run:$ sbgrid-list deepmainmast -
Installation
Use the following command to install this title with the CLI client:$ sbgrid-cli install deepmainmastAvailable operating systems: Linux 64 -
Primary Citation*
G. Terashi, X. Wang, D. Prasad, T. Nakamura, and D. Kihara. 2024. DeepMainmast: integrated protocol of protein structure modeling for cryo-EM with deep learning and structure prediction. Nature Methods. 21, 122-131.
-
*Full citation information available through
-
-
Webinars
SBGrid webinars are hosted with partial support from the NIH R25 Continuing Education for Structural Biology Mentors #GM151273, in collaboration with Co-PI Jamaine Davis of Meharry Medical College.
Topic: Structure modeling and validation using AI-based methods: DeepMainmast and DAQ
Presenter: Genki Terashi, Ph.D. Assistant Research Scientist, Daisuke Kihara's Laboratory, Purdue University
Host: Jason Key
Recorded on March 11, 2025
For more information on deepmainmast & daq:
https://sbgrid.org/software/titles/deepmainmast
https://sbgrid.org/software/titles/daq
-
Default Versions
Linux 64: 20240112 (7.7 GB)
-
Developers
Genki Terashi, Daisuke Kihara, Tsukasa Nakamura, Devashish Prasad, Xiao Wang