Supported Applications
DeepFoldRNA
-
Description
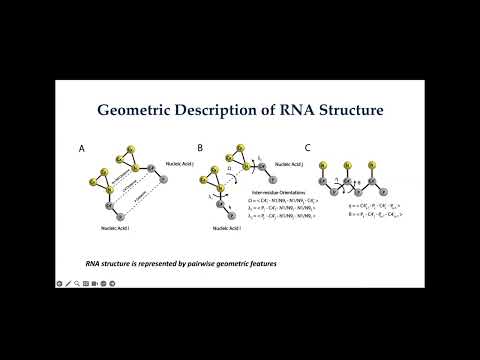
a deep-learning based method for de novo RNA tertiary structure prediction.
-
Usage
To list all executables provided by DeepFoldRNA, run:$ sbgrid-list deepfoldrna -
Installation
Use the following command to install this title with the CLI client:$ sbgrid-cli install deepfoldrnaAvailable operating systems: Linux 64 -
Primary Citation*
R. Pearce, G. S. Omenn, and Y. Zhang. 2022. De Novo RNA Tertiary Structure Prediction at Atomic Resolution Using Geometric Potentials from Deep Learning. bioRxiv 2022.05.15.491755.
-
*Full citation information available through
-
-
Webinars
SBGrid webinars are hosted with partial support from the NIH R25 Continuing Education for Structural Biology Mentors #GM151273, in collaboration with Co-PI Jamaine Davis of Meharry Medical College.
Topic: A deep-learning based method for de novo RNA tertiary structure prediction.
Presenter: Robin Pearce, Ph.D. student, Yang Zhang's lab, University of Michigan Medical School.
Host: Jason Key
Recorded on December 12, 2023
For more information on DeepFoldRNA:
https://sbgrid.org/software/titles/deepfoldrna
https://seq2fun.dcmb.med.umich.edu/DeepFoldRNA/
https://seq2fun.dcmb.med.umich.edu/DeepFoldRNA/help.html
https://zhanggroup.org/forum/
-
Keywords
-
Default Versions
Linux 64: 20220912 (3.4 GB)
-
Developers
Yang Zhang, Robin Pearce, Gilbert S Omenn