Supported Applications
ChimeraX
-
Description
a tool of the next-generation molecular visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. Compared to Chimera, ChimeraX has improved graphics (e.g. interactive ambient shadows); faster handling of large structures (millions of atoms); and a more modern, single-window user interface.
-
Usage
To list all executables provided by ChimeraX, run:$ sbgrid-list chimerax -
Usage Notes
This ChimeraX alpha release is not currently available for CentOS 6 linux.
-
Installation
Use the following command to install this title with the CLI client:$ sbgrid-cli install chimeraxAvailable operating systems: Linux 64, OS X INTEL -
Primary Citation*
T. D. Goddard, C. C. Huang, E. C. Meng, E. F. Pettersen, G. S. Couch, J. H. Morris, and T. E. Ferrin. 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science. 27(1): 14-25.
E. F. Pettersen, T. D. Goddard, C. C. Huang, E. C. Meng, G. S. Couch, T. I. Croll, J. H. Morris, and T. E. Ferrin. 2021. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Science. 30(1): 70-82.
-
*Full citation information available through
-
-
Citation Note
Acknowledge ChimeraX with: "Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases."
You may also cite the ChimeraX home page.
-
Webinars
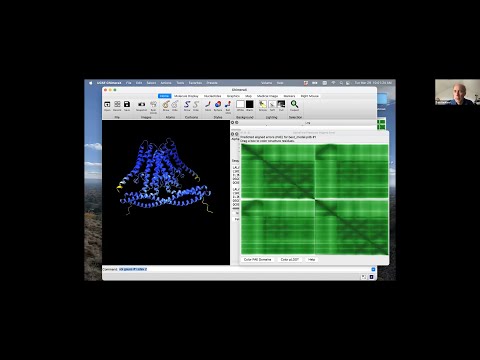
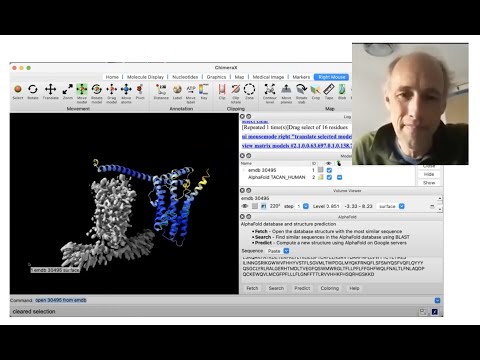
Presenter: Tom Goddard, UCSF Resource for Biocomputing, Visualization, and Informatics - Topic: Using AlphaFold protein structures in ChimeraX for cryoEM modeling
Linked to presented materials: https://www.rbvi.ucsf.edu/chimerax/data/sbgrid-mar2022/alphafold_pae.html
Learn more on the ChimeraX website:
https://www.rbvi.ucsf.edu/chimerax/data/alphafold-nov2021/af_sbgrid.html
This talk was presented as part of the SBGrid Spring Mini-series - Cryo-electron microscopy of membrane proteins: from sample to structure. See the full series lineup at https://sbgrid.org/webinars/
Organized by
Prof. Jamaine Davis, Meharry Medical College
Prof. Piotr Sliz, Harvard Medical School
Prof. Patrick Sexton, ARC Centre for Cryo-electron Microscopy of Membrane Proteins, Monash University
Recorded on March 29, 2022Topic: Using AlphaFold protein structures in ChimeraX for cryoEM modeling
Presenter: Tom Goddard, UCSF Resource for Biocomputing, Visualization, and Informatics
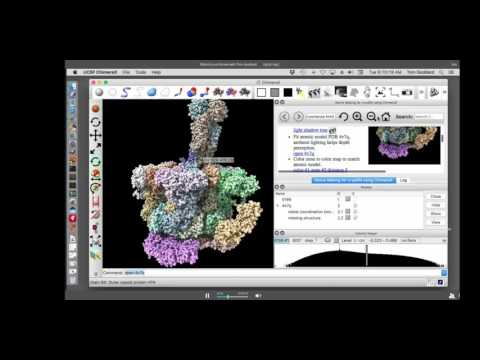
Q&A session also includes Shaun Rawson, who presented during this session on "Effective on-the-fly and downstream processing of CryoEM data." See https://youtu.be/ra2sVFEPtN8
Learn more on the ChimeraX website:
https://www.rbvi.ucsf.edu/chimerax/data/alphafold-nov2021/af_sbgrid.html
This talk was presented as part of the SBGrid Australasian III Mini-series - CryoEM: from Sample to Structure and should appeal to both novice and expert structural biologists.
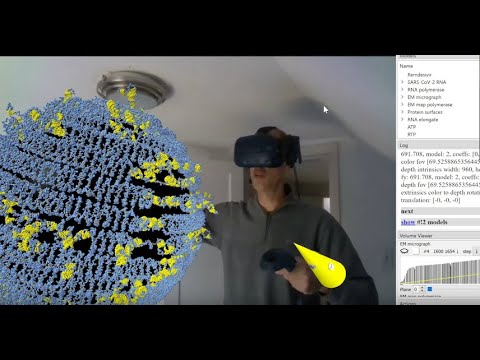
See the full mini-series lineup at https://sbgrid.org/news/sbgrid-university-otago-webinar-series.Topic: Making Augmented Reality Videos with ChimeraX
Presenter: Tom Goddard, University of California, San Francisco, Resource for Biocomputing, Visualization, and Informatics
The Zoom recording of this presentation is pretty choppy. We recommend you also check out the video Tom recorded and posted on his own channel here: https://youtu.be/dKNbRRRFhqY
Slides from talk available at https://www.rbvi.ucsf.edu/chimerax/data/sbgrid-july2020/
Host: Pete Meyer
Recorded on July 7, 2020
-
Keywords
-
Default Versions
Linux 64: 1.11 (7 bytes)
OS X INTEL: 1.11 (2.0 GB) -
Other Versions
Linux 64:
1.10 (10 bytes) , 1.10.1 (9 bytes) , 1.10.1_c8 (1.7 GB) , 1.10.1_c9 (1.6 GB) , 1.10-rc (13 bytes) , 1.10-rc_rhel8 (1.5 GB) , 1.10-rc_rhel9 (1.7 GB) , 1.10_rhel8 (1.8 GB) , 1.10_rhel9 (1.7 GB) , 1.11_c8 (1.9 GB) , 1.11_c9 (2.2 GB) , 1.3 (1.5 GB) , 1.3_c8 (1.5 GB) , 1.3_scipion (1.3 GB) , 1.5 (6 bytes) , 1.5_c8 (1.4 GB) , 1.5_c9 (1.4 GB) , 1.6.1 (9 bytes) , 1.6.1_c8 (1.7 GB) , 1.6.1_c9 (1.6 GB) , 1.9 (9 bytes) , 1.9_rhel8 (2.1 GB) , 1.9_rhel9 (2.1 GB) , 20250305 (11 bytes) , 20250305_c8 (1.8 GB) , 20250305_c9 (1.8 GB) , 20251126 (11 bytes) , 20251126_c8 (1.7 GB) , 20251126_c9 (1.7 GB) , 20251214 (11 bytes) , 20251214_c10 (1.7 GB) , 20251214_c8 (1.8 GB) , 20251214_c9 (1.7 GB) , 20260215 (11 bytes) , 20260215_c10 (0 bytes) , 20260215_c9 (1.8 GB) -
OS X INTEL:
1.10 (1.9 GB) , 1.10.1 (1.9 GB) , 1.10-rc (1.9 GB) , 1.4 (1.1 GB) , 1.6.1 (1.6 GB) , 1.7.1 (1.2 GB) , 1.8 (1.9 GB) , 1.9 (1.9 GB) , 20230908 (1.6 GB) , 20250305 (1.9 GB) , 20250314 (1.9 GB) , 20251016-arm64 (1.5 GB) , 20260215 (2.4 GB)
Developers
Conrad Huang, Elaine Meng, Eric Pettersen, Greg Couch, John Morris, Thomas Ferrin, Thomas Goddard