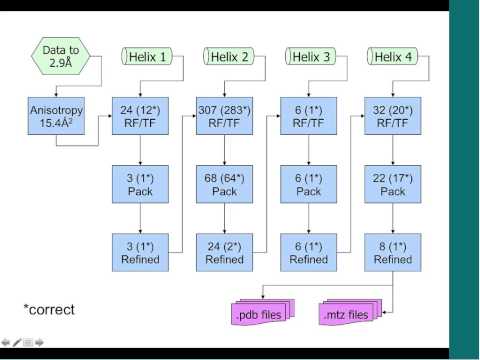
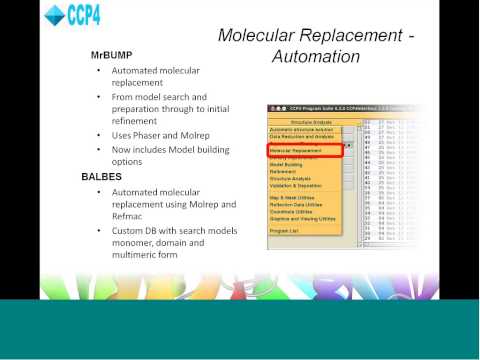
Unstructured
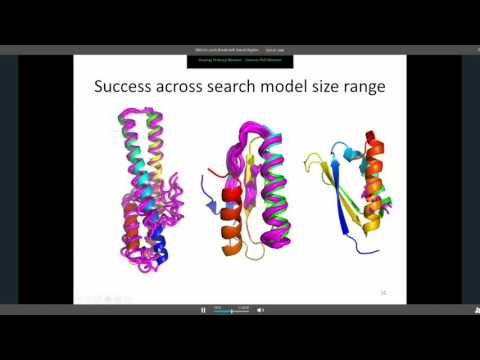
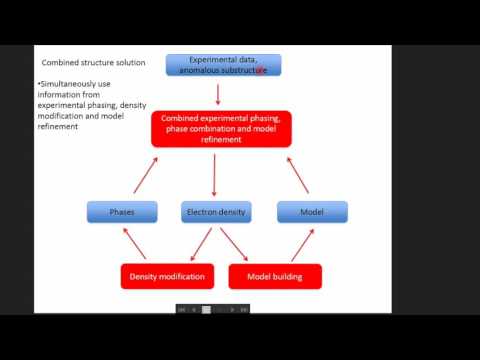
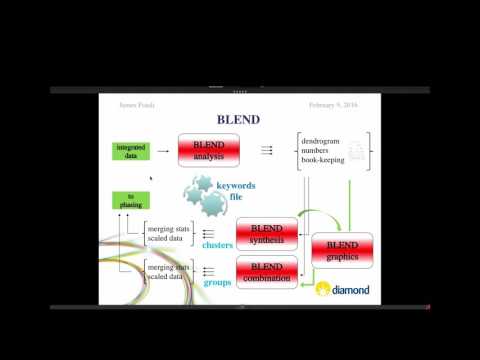
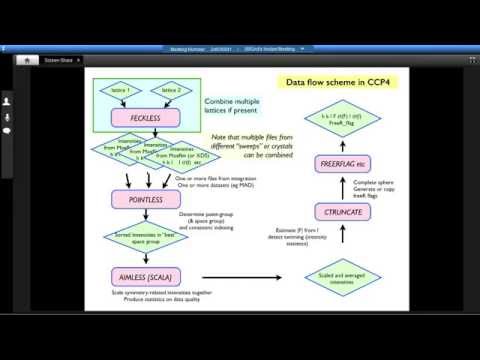
Sit down in front of a newly installed copy of CCP4 today, and you will find approximately 250 computer programs for solving protein structures. The list of programs includes several with catchy names, such as beast (for molecular replacement), dimple (for ligand identification in difference maps), crank (for experimental phasing) and buccaneer (for model building), and some cryptic, such as seqwt and npo. Nearly two dozen applications support file manipulations and format conversions. Still more are riders-on, either deprecated or unsupported.
The seeming mishmash is so by design. "CCP4 has always been a very loose collaboration," says Phil Evans, a structural biologist at the Medical Research Council Laboratory of Molecular Biology in Cambridge, UK, and one of the earliest members of the team that created CCP4 in 1979. …
Read the full story here.