Supported Applications
Amber
-
Description
a suite of biomolecular simulation programs. It began in the late 1970's, and is maintained by an active development community.
-
Usage
To list all executables provided by Amber, run:$ sbgrid-list amber -
Installation
Use the following command to install this title with the CLI client:$ sbgrid-cli install amberAvailable operating systems: Linux 64 -
Webinars
SBGrid webinars are hosted with partial support from the NIH R25 Continuing Education for Structural Biology Mentors #GM151273, in collaboration with Co-PI Jamaine Davis of Meharry Medical College.
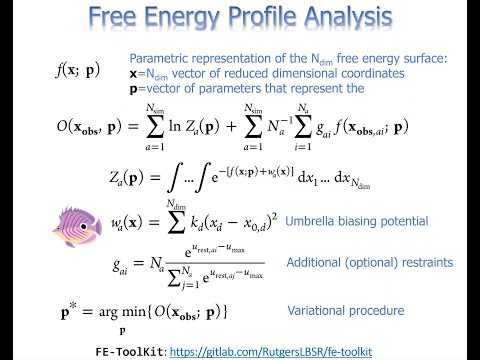
Topic: Free energy methods in Amber.
Presenter: Professor Darrin York, Rutgers University.
Host: Jason Key
Recorded on May 14, 2024
For more information on Amber:
https://sbgrid.org/software/titles/amber
https://theory.rutgers.edu/
-
Keywords
-
Default Versions
Linux 64: 24 (3.1 GB)
-
Other Versions
Linux 64:
22 (5.4 GB)