The Language of Gene Regulation
Daniel Panne
European Molecular Biology Laboratory, Grenoble
Published November 21, 2017
To find an analogy for his studies of how genes are turned on and off, Daniel Panne turns to the prehistoric drawings he viewed on a recent family vacation. The famous Lascaux Cave is a half-day’s drive from Grenoble, France, where he has headed a research group for nine years at the European Molecular Biology Laboratory (EMBL). Scenes of horses, stags, and other images depicted on cave walls and ceilings showcase some of the earliest examples of human art and thought.
“Some linguists consider such examples of abstract symbolic reasoning as an indication of the ability to produce language, which seems to have arisen in humans only around 100,000 years ago,” Panne says. By some theories, language is generated by combinatorial usage of a limited repertoire of words strung together in meaningful ways. The mixing and matching of words and phrases is called a “merge” operation in linguist jargon. This architecture of language is conceptually similar to the molecular language that regulates gene expression, Panne says.
To respond to a cell’s need at any given time, a limited number of proteins touch down on the chromatin packaging of DNA to unlock a gene or change its activity. The proteins, called transcription factors, can combine in nearly limitless ways to target different genes, generating a rich regulatory syntax. As in the human language analogy, the critical feature is the merge operation. Transcription factors come together like words in a sentence to jointly regulate gene expression, Panne says. The transcription factors merge onto common scaffold proteins, usually CBP or its look-alike p300, called co-activators.
On an evolutionary note, co-activators CBP and p300 are found only in multicellular organisms, and not more simple one-celled life forms. The fossil record suggests complex multicellular life emerged about 600 million years ago in a relatively short period termed the “Cambrian explosion,” during which most major animal phyla appeared. Panne wonders if that pivotal moment on Earth may have gone hand in hand with the unique merge function of transcription factors and a dramatically changed ability to regulate genome expression.
To understand gene regulation, Panne works on a cell-free model of the human interferon beta response to viral infection. Panne began working on the model as a postdoctoral fellow in the laboratory of Steve Harrison at Harvard Medical School. The model system was first developed in the Harvard lab of Tom Maniatis.
“This is the only system I know about that is amenable for structural work on how the cellular signal comes down to the chromatin and DNA and that can provide an answer about how a signal activates a eukaryotic gene,” Panne says. Interferon signaling is central to the innate immune system, so answers to these questions also can lead to better understanding of cancers and diseases and new drugs related to immune activity or malfunction.
A virus added to the model system triggers a well-characterized and synchronized response. Beginning with the pathogen pattern recognition receptors (PPRRs), the signal travels like clockwork through various molecular components and culminates in interferon beta gene regulation. The complicated gene targeting machinery works at the level of the chromatin packaging of DNA. The chromatin must be unpacked to expose genes for transcription and then put back together to shut them down.
In the Harrison laboratory, Panne determined the structure of an “enhanceosome,” a term that refers to a collection of transcription factors that binds to a DNA element and regulates gene expression. Panne continued to refine the system for structural work after moving to EMBL in 2007. His group determined active and inactive structures of an upstream kinase (Tank-binding kinase I (TBKI)) important in interferon beta regulation, published in 2013 in Cell Reports. Previously, TBK1 had been implicated in uncontrolled rheumatoid arthritis, obesity-related metabolic disease, and certain cancers. Panne’s findings led to a side project on structure-based drug design with industry partners to develop novel drugs.
A major part of the Panne lab focuses on chromatin and its role in gene regulation. Chromatin refers to the DNA packaging needed to squeeze the long strands into each cell. For each chromosome, DNA is spooled around histones. Four histones make a nucleosome, and a string of nucleosomes make up the chromatin. Panne’s group investigates how chromatin is assembled and disassembled, how it is modified, and what that all means for gene regulation.
After DNA is copied, transcribed, or repaired, its chromatin packaging needs to be reassembled. In a biochemical study in 2017 in eLife, Panne’s group reported on the first step of nucleosome assembly. In the first step, an enzyme called CAF1 binds to the newly made DNA and deposits two histone copies. In a second study, published in 2016 in the EMBO Journal, the Panne group reported on the second nucleosome assembly step. Here, a protein called Nap1 catalyzes the second set of histones. The four histones create a new nucleosome spool for that section of DNA.
Next comes the disassembly problem. To access a gene, the transcription machinery must get past the chromatin packaging of DNA. For this, a group of transcription factors and their co-activator land on the genetic package and deploy a chemical tag to flag the relevant chromatin. In the interferon beta system, the transcription factors, or “enhanceosome,” lands on a stretch of DNA between nucleosomes. Then p300 modifies nearby chromatin with a chemical tag called acetylation, somehow releasing the inhibitory chromatin structure to activate a gene.
In cancer research, the CBP and p300 proteins were known as tumor suppressors until the Panne lab showed that several known cancer mutations could cause p300 to over-activate itself. They reported the protein structure in Nature Structural and Molecular Biology in 2013. Recently, other researchers found p300 had more tricks up its sleeve. It could make other modifications to chromatin in response to being starved or well-nourished. In Nature Chemical Biology in 2017, Panne’s group reported how that works. In well-nourished cells, p300 uses an abundant supply of Acetyl-CoA to tag chromatin with the acetyl group and change gene regulation. In starved cells, on the other hand, p300 deploys chemically similar CoA variants. The work connects cell metabolism to gene regulation.
Panne’s research addresses an area of controversy that permeates the field of epigenetics and erupted publicly after a 2016 New Yorker article on the topic. Epigenetics refers to factors that change gene behavior without altering the DNA sequence. The article claimed the phenomenon was primarily driven by epigenetic modification of chromatin. The idea is that small chemical tags, such as acetylation, on chromatin marking active or inactive genes would self-propagate independently of the underlying DNA sequence and direct the activity of the genome. The idea arose from findings that many chromatin modifiers, such as p300, contain both domains that catalyze chromatin modification and, importantly, other domains that read the chromatin modification status (see accompanying p300 image).
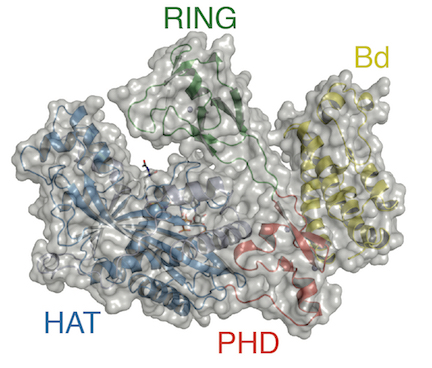
In contrast, many scientists consider that chromatin does not form an independent epigenetic layer of the genome and that chromatin modifiers do not operate independently of a DNA sequence specific targeting mechanism, such as transcription factors. “What the New Yorker article gets wrong is subtle but important,” Panne says. “The article says modified chromatin dictates the biology, as if the tail wags the dog. But it turns out that modified chromatin is the result of signaling that targets the DNA.”
In other words, modification of chromatin arises due to cellular signaling to particular genome loci and does not appear to be propagated independently of genome targeting. It will be important, Panne says, to dissect in detail how scaffold proteins, such as the CBP and p300 co-activators, contribute to the rich gene regulatory language, how such chromatin modifiers are targeted to the genome, how their activity is regulated, and how chromatin modifications contribute to the signaling reaction.
-Carol Cruzan Morton





































































































































