Intron Intrigue
Navtej Toor
University of California at San Diego
Published February 20, 2015
When Navtej Toor started searching for a post-doc in 2004, he applied to just one lab, that of Anna Pyle at Yale University. It was a long distance from the University of Calgary, where he’d done his undergraduate and graduate studies in biochemistry, and a far cry from his nearby rural hometown of Sparwood, British Columbia. But Pyle’s interests most closely resembled his own.
Toor, now an assistant professor of chemistry and biochemistry at the University of California, San Diego, had become interested in RNA as an undergrad. Later, when he saw the high-resolution structures of the large and small ribosomal units, published in 2000, he became interested in structural biology. “With structural biology, they explained years of biochemistry in a single experiment,” says Toor.
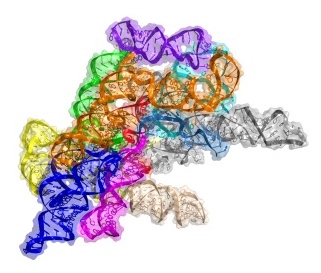
For his post-doc, Toor wanted to solve the crystal structure of a group II intron RNA, a molecule formed from a non-coding portion of DNA that delineates genes in lower level organisms such as bacteria. In humans, introns punctuate genes and support, with the help of a large molecular machine called the spliceosome, a process called splicing that allows one gene to code for multiple products.
But Pyle’s lab was not a crystallography lab, Toor was not a crystallographer, and on top of that, earlier attempts to solve this structure had not succeeded. This inauspicious triad, however, was the essence of Toor’s strategy. Instead of joining a crystallography lab that might not be willing to let him fail, he joined a lab with intron expertise, and brought with him enthusiasm and a plan. He even found SBGrid, which Pyle’s lab joined in a hurry, to help him get all the software he needed installed and running.
The appeal of examining these tiny parts of the gene transcription machinery is many-layered. Approximately fifteen percent of human genetic diseases result from gene splicing errors. But the mechanics of human gene splicing is complex. The spliceosome itself is made of hundreds of proteins. The core of its machinery grabs onto introns, cuts them out, and knits genes back together to begin the protein translation process.
But in bacteria, introns cut themselves out and stitch the remains back together. “The core of the spliceosome is thought to be very similar to the core of group II introns, so we’re studying them to try to get insights into how the spliceosome works,” says Toor.
After four years of work, including many alterations to the solvent-exposed residues on the RNA to get usable crystals of group II introns, Toor succeeded in solving the structure of a bacterial group II intron in 2008. It was an exciting accomplishment, but the intron was very primitive. “It was missing a lot of the machinery you need for splicing,” says Toor.
For example, this intron used simple hydrolysis as a splicing mechanism, but other group II introns — and introns in humans — form a lasso at the splice site. “This lariat is conserved all the way from bacteria to humans,” says Toor.
While the lariat is clearly important for splicing, Toor believes that it is also required for splicing to happen in reverse. Reverse splicing was necessary for the genome to evolve from a sparse population of introns in bacteria to abundant numbers in humans. Introns make up about 25 percent of the human genome. “They were not always there,” says Toor. “They spread from an initial ancestor, but how? It’s an understudied part of biology, looking at the evolution of non-coding regions.”
After moving to UCSD in 2009, Toor’s lab solved the structure of a group II intron lariat from a form of algae. The structure, published in 2014, provides a snapshot of the end state of the splicing process, so Toor is now focused on solving earlier stages. “By capturing the entire movie of the lariat-forming intron, we hope to be able to understand how introns reverse splice and spread throughout genomes,” he says.
X-ray crystallography is still Toor’s primary tool, but he has recently begun to use electron microscopy in the hopes that it might provide insights without requiring him to form crystals. “If we can bypass that, it will save us a lot of time,” says Toor.
-- Elizabeth Dougherty





































































































































